REVIEW ARTICLE OPEN ACCESS
Organoids as Next-Generation Models of Tumor Drug Resistance
Ling Wang1,2,3*, Qin Tang1,2,3*, Lin Zhong1,2,3*, Misi He1,2,3, Xueping Zhu1,2,3, Qian Zheng1,2,3, Ling Long1,2,3, Ya Wang1,2,3, Qingxiu Jiang1,2,3, Haixia Wang1,2,3 #, Dongling Zou1,2,3#
Received 2025 Sept 4
Accepted 2025 Nov 16
Epub ahead of print: December 2025
Published in issue 2026 Feb 15
Correspondence: Dongling Zou - Email: zoudl_cqch@cqu.edu.cn
Haixia Wang - Email: wanghx1985@126.com
The author’s information is available at the end of the article.
© 2026 The Author(s). Published by GCINC Press. Open Access licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. To view a copy: https://creativecommons.org/licenses/by/4.0/
Abstract
Tumor drug resistance is a major clinical challenge and critical to patient outcomes. Although extensive studies on mechanisms and drug development using cell and animal models exist, clinical results remain limited or ineffective. Recently, organoids have emerged as a powerful model that closely reproduces the structural and genetic features of tumors in vivo, making them an increasingly valuable platform for investigating resistance mechanisms and developing new therapeutic strategies. We reviewed organoid applications in elucidating tumor drug-resistance mechanisms, advancing therapies, and supporting clinical research, underscoring their critical role in overcoming treatment resistance. These mechanisms include abnormal cancer-related signaling, interactions between tumor and microenvironmental cells, DNA repair, epigenetic changes, and modifications in tumor cell membrane proteins. As an emerging preclinical model, organoids help bridge the gap between basic research and clinical practice, supporting the development of strategies to reverse drug resistance. These strategies include designing new targeted drugs, testing the efficacy of combination therapies, repurposing existing drugs, and screening personalized treatments for rare tumors. Moreover, in clinical trials, organoids can help guide therapy selection and enable preclinical evaluation of emerging therapeutic strategies. Although organoids lack the full in vivo microenvironment and an intact vascular system, advances in co-culture platforms and microfluidic chip technologies are steadily overcoming these limitations. As organoids integrate with such innovations, their role in drug-resistance research will continue to grow.
Keywords: Organoids, Drug resistance, 3D cultures, Microfluidic chip technologies, Preclinical models, Tumor microenvironment, Personalized medicine
1. Introduction
Tumor drug resistance remains one of the major challenges for anti-tumor therapeutic strategies (1,2). The widespread use of numerous novel agents in clinical practice has driven the development of multidrug resistance, posing significant challenges for subsequent treatment options. Multiple studies revealed that tumor drug resistance is based upon a plethora of distinct mechanisms (3). Studies based on cell and animal experiments are insufficient to address the diverse drug-resistance profiles observed in clinical patients. The emergence of organoids has led to a new era for research on drug resistance. The organoids simulate tumor heterogeneity and retain the parent tumor's molecular expression patterns, making them an ideal platform for studying tumor heterogeneity and drug resistance mechanisms (4-6).
In recent years, as organoid technology has advanced, its application to drug resistance research has attracted growing attention. Due to the high fidelity of organoids, patient-specific organoid biobanks can simulate clinical treatment pressures, dynamically track the evolution of drug resistance, and support in-depth analyses of the molecular mechanisms of drug resistance (7). Furthermore, the combination of genetic engineering and microfluidic technology with organoid models has expanded their potential for personalized treatment strategies (8). The integration of novel technologies has enhanced the applicability of biological models, providing a powerful supporting mechanism to precision medicine (9). Multiple studies in 2025 demonstrated that organoid-based drug resistance models made breakthrough advances in solid tumors (10,11), including lung cancer, gynecological malignancies, colorectal cancer, and pancreatic cancer (PC) (12-16). However, the inability of organoids to replicate the tumor microenvironment (TME) and vascular network limits their wide application in cancer research (17). The combination of emerging technologies (e.g., 3D bioprinting, organoid-on-chips, microfluidic devices) with organoids is being used to create novel engineered models that more accurately replicate organogenesis, physiological processes, and disease progression (18,19), thus providing new perspectives for drug resistance research (Figure 1).
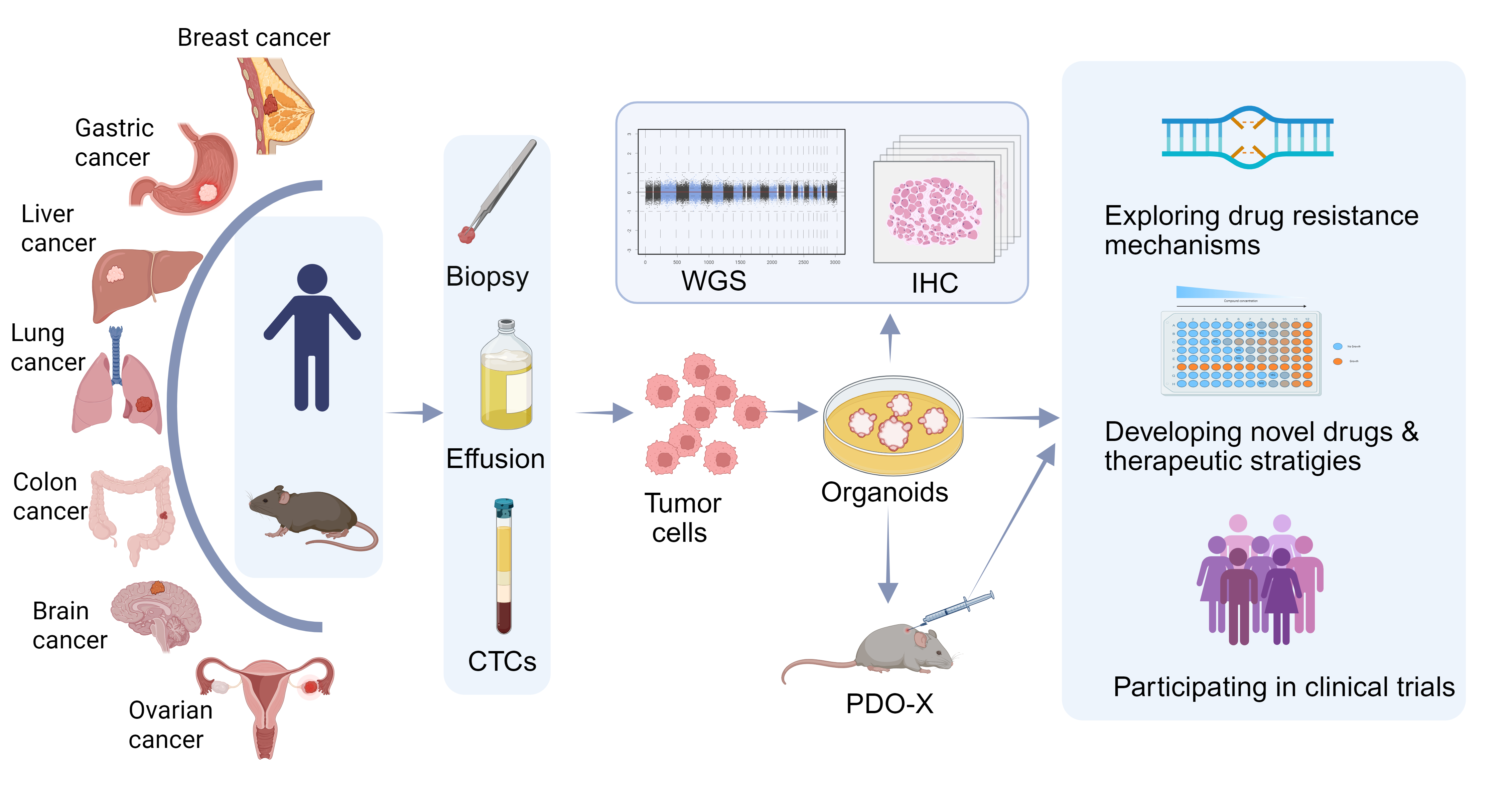
Figure 1. Overview of Organoid Construction and Its Application in Tumor Drug Resistance. Tumor tissues, including biopsy samples, malignant effusions, and circulating tumor cells (CTCs), are collected from humans and animals to extract tumor cells for organoid cultivation. The consistency between organoids and the original tumors is validated through immunohistochemistry (IHC) and whole-genome sequencing (WGS). Organoids or animal models transplanted with organoids (PDO-X) can be applied to explore drug resistance mechanisms, develop new drugs and therapeutic strategies, and participate in clinical trials. Figure created using BioRender.
This review systematically outlines the applications of organoids in drug-resistance research, including their use to uncover resistance mechanisms and guide the development of new targeted agents and therapeutic strategies. In addition, organoids have increasingly been incorporated into clinical trials, where they play multiple roles in studies related to drug resistance. We aim to provide new perspectives to promote the application of organoids in research on drug resistance and to establish a theoretical framework for precision medicine research.
2. The Role of Organoids in Tumor Drug Resistance Mechanisms
2.1 Abnormal cancer-associated signaling
2.1.1 Identifying resistance-associated signaling
Organoids provide robust in vitro platforms for elucidating the mechanisms underlying tumor drug resistance (Fig. 2). For example, MEX3A and Bcl-xl have been identified as key mediators of chemoresistant relapse in colorectal cancer and cholangiocarcinoma organoids, respectively (20,21). In the co-culture system of gastric cancer patient-derived organoids (PDOs) and macrophages, aurora kinase inhibitors (AURKi) can induce a senescent phenotype, characterized by increased cell diameter, strong senescence-associated β-galactosidase staining, and multinucleated giant cells. The senescent cancer cells secrete large amounts of the chemokine MCP-1/CCL2, which recruits and induces macrophages to an M2-type polarization, resulting in attenuated innate immune responses and reduced killing of cancer cells (22). Using organoids derived from circulating tumor cells (CTCs) and pleural effusion, Würth et al. identified NRG1-HER3 signaling as a key pathway required for cancer dissemination. The dynamic interplay between NRG1-HER3 and FGFR1 signaling mediates cancer cell plasticity, leading to therapeutic resistance (23). Additionally, organoid models can also simulate adaptive changes under drug pressure to reveal the evolution of resistance. Using single-cell sequencing of lung cancer organoids, Tong et al. revealed that tumors activate a lineage plasticity program, adeno-to-squamous transition (AST), that mediates KRAS inhibitor resistance (24). Under the exposure of cisplatin, tongue cancer cells of the chemo-resistant tongue cancer organoids (TCOs) reconstituted organoids after cisplatin withdrawal, while those of chemo-sensitive TCOs did not (25).
Comparing drug-resistant and drug-sensitive models is a classic approach for identifying resistance-related signaling. Organoids provide a good preclinical model, capable of mapping drug resistance and sensitivity profiles in rare tumors and identifying effective therapeutic targets (26). Docetaxel-sensitive and -resistant gastric cancer organoids were developed to explore resistance mechanisms using organoid-based single-cell RNA sequencing. In resistant organoids, the secretory, immune-chemotactic, and transitional gastric cancer cell populations were increased. Resistance-related genes were upregulated, regulating ROS production, autophagy, and apoptosis (27) (Fig. 2A). Exploration of drug resistance-related genes and pathways can lead to the development of novel therapeutic targets for overcoming drug resistance and improving treatment outcomes for cancer patients. Sase et al. established a library of TCOs, which were divided into chemo-resistant and chemo-sensitive groups. Comparative analysis of TCOs revealed that “heritable” embryonic diapause-like features are involved in the development of chemo-resistance. Mechanistically, the activation of the autophagy and cholesterol biosynthetic pathways confers TCOs chemo-resistance. Inhibitors of the two pathways could convert chemo-resistant TCOs to chemo-sensitive TCOs and almost completely kill chemo-resistant TCOs (25).
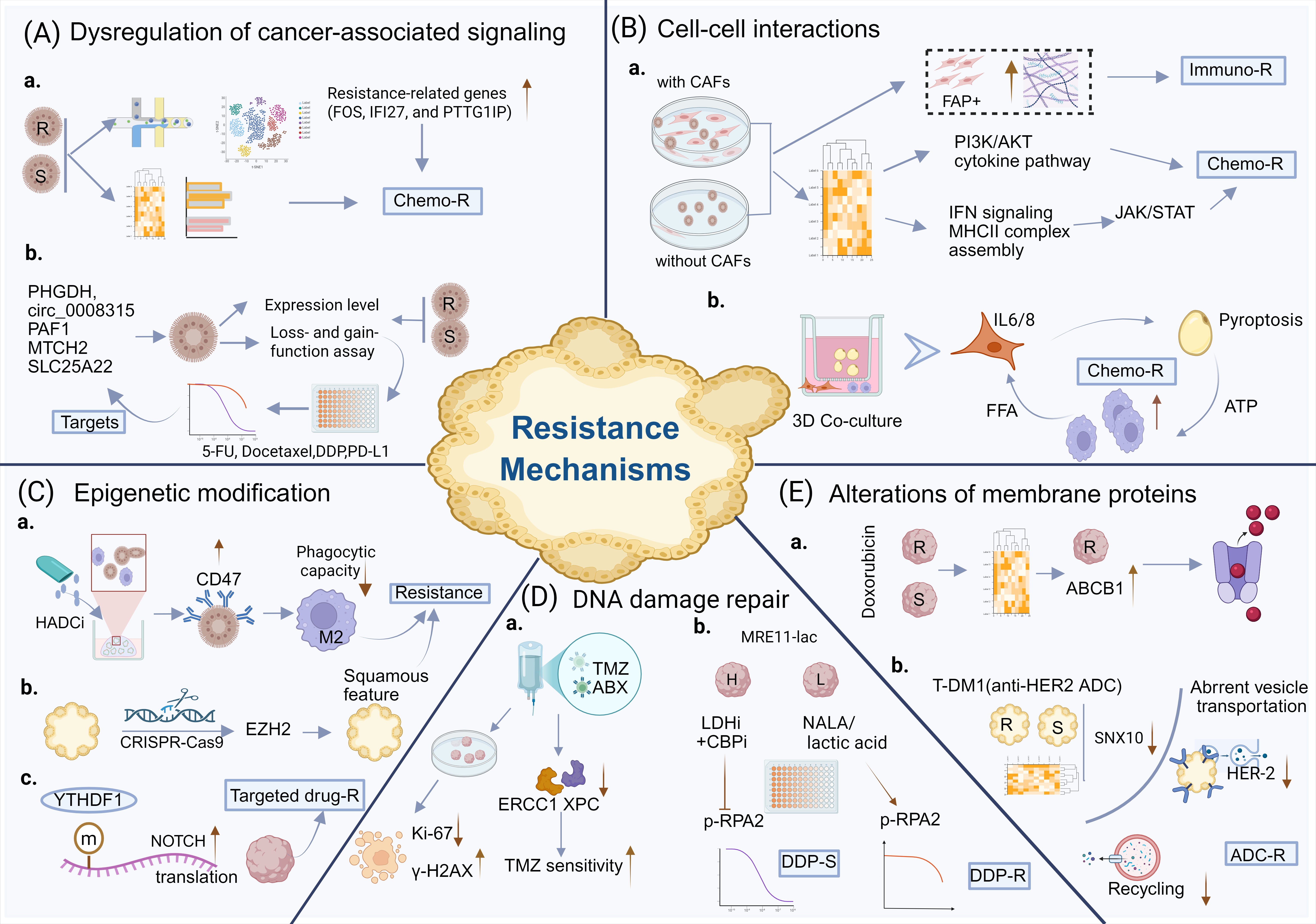
Figure 2. The roles of organoids in research on drug resistance mechanisms. (A). Organoids can be utilized to explore abnormal tumor-related signaling and validate potential targets. Drug-resistant and drug-sensitive organoids are analyzed using single-cell sequencing or multi-omics approaches to identify resistance-associated genetic and signaling pathways. Furthermore, loss- and gain-of-function assays and drug screening in organoids can be used to validate potential targets. (B). Co-culture systems of tumor organoids with microenvironment components, such as CAFs, macrophages, and adipocytes, help elucidate the role of cell-microenvironment interactions in drug resistance. (C). Organoids also play a significant role in investigating epigenetic dysregulation. Organoids combined with CRISPR-Cas9 gene-editing technology serve as a powerful tool for exploring mechanisms of drug resistance. (D). Elevated expression of ABCB1 in drug-resistant organoids promotes drug efflux, leading to drug resistance. Organoids provide a model for studying ADC resistance. (E). The Organoids contribute to the investigation and validation of DNA damage repair in drug-resistant (R) and drug-sensitive (S) cells. H, high level of MRE11 lactylation. L, low level of MRE11 lactylation. TMZ, temozolomide. ABX, albumin-bound paclitaxel. HADCi, histone deacetylase inhibitor. LDHi, lactate dehydrogenase inhibitor. The brown arrow indicates changes in gene expression levels or drug sensitivity, while the blue arrow indicates the directionality of the schematic diagram. Figure created using BioRender.
2.1.2 Validating drug resistance-related targets
Organoids serve as powerful systems for validating proposed resistance mechanisms (Fig. 2A). For instance, organoids with elevated PHGDH expression displayed 5-FU resistance through Hedgehog signaling activation. Using CRC organoid models, a combined 5-FU and Hedgehog inhibitor strategy was subsequently shown to be effective in overcoming this resistance (28). Additionally, organoids are considered robust in vitro 3D models for functional studies that demonstrate the significance of drug-resistance-related targets. Loss- and gain-of-function experiments were performed on breast organoids, elucidating anticancer activity through MTCH2 inhibition (29). Similarly, PAF1 depletion was performed in prostate cancer organoids, demonstrating PAF1's regulatory function in docetaxel resistance (30). The upregulation of circ_0008315 was validated in cisplatin-resistant gastric cancer organoids, and its knockdown improved therapeutic efficacy and reversed cisplatin resistance (31). Notably, T-cell-incorporated organoid models validated SLC25A22, a glutamine metabolism transporter, as a key driver of elevated glutamine utilization, thereby reversing the immunosuppressive microenvironment and enhancing sensitivity to anti-PD-1 therapy (32).
2.2 Cell-cell interactions
Due to the complex cell-cell interactions in the TME, the mechanisms of drug resistance are highly intricate (33). The microenvironment comprises cancer-associated fibroblasts (CAFs), adipocytes, endothelial cells, and immune cells, all of which are crucial for drug resistance (34,35). Co-culture systems combining organoids with components of the TME provide powerful platforms for modeling interactions between cancer cells and surrounding stromal and immune cells (36-38). For example, a cancer organoid–stroma biobank enables detailed molecular subtype characterization, supports individualized preclinical testing, and helps identify stromal-driven resistance mechanisms (39).
2.2.1 Nonimmune cells and cancer cells
Co-culture models integrating organoids with cancer-associated fibroblasts (CAFs) provide a robust 3D in vitro platform for mechanistic validation. In a 3D co-culture system using human ovarian cancer organoids, CAFs were shown to promote organoid growth and confer chemoprotection by activating the PI3K–AKT pathway and cytokine-mediated signaling (40). The spatial co-enrichment of CAFs with positive fibroblast activation protein (FAP+) and collagen in urachal cancer PDOs leads to stromal-mediated immune evasion and resistance to immunotherapy (41). A co-culture system of CRC organoids with CAFs revealed that chemoresistance-associated interferon-α/β signaling and MHC class II complex assembly contribute to drug resistance by activating the JAK/STAT pathway (42) (Fig. 2B). Endothelial cells, as key components of the cellular niche, can support organoids during long-term culture. They also modulate the expression profiles of liver organoids—particularly genes involved in extracellular matrix organization and receptor-mediated signaling, through paracrine interactions (43). A three-cell co-culture model incorporating ovarian cancer (OC) cells, adipocytes, and macrophages was developed to mimic the omental tumor microenvironment. In this system, IL-6 and IL-8 secreted by OC cells induced pyroptosis in omental adipocytes. The resulting pyroptotic adipocytes released ATP, which promoted macrophage infiltration, and liberated free fatty acids (FFAs) into the microenvironment. These FFAs were subsequently taken up by OC cells, ultimately enhancing chemoresistance (44) (Fig. 2B).
2.2.2 Immune cells and cancer cells
Jiang et al. used a droplet-based microfluidic platform to generate PDOs and examine the interaction between macrophages and PC cells via co-culture of PC organoids (PCOs) and macrophages. In this system, macrophage-derived CCL5 activated the CCR5/AKT/Sp1/CD44 signaling pathway, promoting stemness and chemoresistance in PC cells, while PC cell-derived AREG stimulated CCL5 production in macrophages via the Hippo–YAP pathway. Combining gemcitabine with agents targeting this feedback loop demonstrated promising therapeutic potential (45). In co-culture assays of CRC organoids and T cells, USP19 was identified as a key factor in immune evasion. Targeting USP19, along with ɑPD-L1 therapy, reduced PD-L1 levels and boosted CD8 T-cell activation and GZMB infiltration, leading to strong anti-tumor effects (46). Similarly, co-culturing lung cancer organoids with peripheral blood lymphocytes enables the study of tumor-T cell interactions and may help identify, in a personalized way, factors that influence immunotherapy sensitivity or resistance (47). Organoid-based co-culture systems also aim to explore the complex communication between cancer cells and dendritic cells (48).
In addition, the Air-Liquid Interface Three-Dimensional (ALI-3D) culture system can preserve native stromal populations, including fibroblasts, macrophages, and lymphocytes (49). Based on this platform, the PDOs' response to anti-PD-1 in vitro was comparable to that observed in clinical trials (50). In the ALI-3D co-culture of mouse PC cells and bone marrow-derived dendritic cells, dynamic glycolytic reprogramming inhibited dendritic cell antigen presentation and mitochondrial function, resulting in poor immunotherapeutic efficacy (51). Recently, microfluidic tumor organoid-on-a-chip platforms have been shown to recapitulate key features of the TME, and the immune contexture has emerged as an innovative, reliable tool to investigate the role of the TME in counteracting antitumor immunity and the mechanisms of immunotherapy resistance (52,53). The microfluidic platform can address the critical need for multicellular tumor models, potentially yielding novel biological insights into cell-cell crosstalk (54). Reprogramming the interactions between cancer cells and TME components may open a new therapeutic avenue in oncology and improve clinical translation.
2.3 The alteration of epigenetic modification
Epigenetic modifications trigger diverse biological processes that can modify cellular phenotypes (55-57), including disease progression and resistance (58, 59). Human intestinal epithelial organoids retain the location-specific transcriptional and epigenetic profiles of the intestinal segment from which they were derived, even after long-term culture (60). Growing evidence indicates that organoids serve as powerful disease-specific models for epigenetic research (61,62). PDOs not only serve as basic research models for RNA modification in cancer therapeutic resistance but also as a preclinical platform to evaluate the clinical benefits of small-molecule modulators targeting RNA modifications (63,64). For example, YTHDF1 was found to promote stemness and aberrant activation of NOTCH signaling in hepatocellular carcinoma (HCC) PDOs, thereby driving resistance to lenvatinib and sorafenib (65) (Fig. 2C). NAT10, which mediates RNA N4-acetylcytidine (ac4c) modification, stabilized AHNAK mRNA by protecting it from exonucleases. AHNAK-dependent DNA damage repair (DDR) is essential for NAT10-driven cisplatin resistance. Notably, Remodelin (an inhibitor of NAT10) sensitizes bladder cancer organoids to cisplatin, suggesting that NAT10 may be a therapeutic target to overcome cisplatin resistance (66). Xu et al. established a co-culture system of CRC organoids and primary human macrophages to reveal the mechanisms of HDAC inhibitor (HDACi) resistance. HDACi treatment epigenetically upregulated CD47 in CRC organoids, thereby polarizing macrophages toward a pro-tumor M2 phenotype and impairing macrophage phagocytic capacity against tumor cells (67) (Fig. 2C). A CRISPR–Cas9 loss-of-function screen in gastric cancer organoids identified EZH2 as a critical epigenetic regulator of drug resistance. Loss of EZH2 induced squamous-like features in the organoids and conferred increased chemoresistance (68) (Fig. 2C).
2.4 DNA damage repair
DNA damage repair represents a critical factor in the development of resistance to chemotherapy and targeted therapy (69, 70). Recently, organoids have been used to investigate DNA damage repair and potential drug-resistance targets (71-73). Long-term use of temozolomide (TMZ) can lead to decreased sensitivity and even chemoresistance in glioblastoma multiforme (GBM). In contrast, albumin-bound paclitaxel (ABX) could enhance the efficacy of TMZ by increasing DNA damage by disrupting the expression and nuclear translocation of DNA repair proteins ERCC1 and XPC. The combinatorial regimen was applied in GBM organoids, which exhibited reduced organoid diameter, reduced Ki-67 expression, and increased γ-H2AX compared with TMZ monotherapy (74) (Fig. 2D). In addition, lactate was shown to promote resistance in gastric cancer PDOs to multiple DNA-damaging therapies, including cisplatin, etoposide, adriamycin, and ionizing radiation, by enhancing homologous recombination (HR)-mediated DNA repair (75). Mechanistically, lactylation of MRE11, a key HR protein, facilitated DNA end resection and promoted HR. CRC PDOs were subsequently stratified into high and low MRE11 K673-lactylation groups to delineate how this modification contributes to resistance to cisplatin and PARP inhibitors (76) (Fig. 2D). Polθ, which is responsible for the interaction, annealing, and extension of short single-stranded (ss), initiates DNA synthesis to fill in the gaps (77, 78). Combined defects in BRCA1 and the Shieldin complex have been shown to confer PARPi resistance. A PI3K inhibitor selectively sensitized BRCA1-mutant breast cancer PDOs, but not BRCA1-wild-type PDOs, thereby overcoming resistance to carboplatin and olaparib (79).
2.5 Alterations of membrane proteins of tumor cells
Reduced intracellular drug accumulation, whether through increased efflux or decreased influx, can partly cause drug resistance or non-responsiveness. Although research on drug resistance mechanisms involving organoids is continuously evolving, there is relatively little research on drug efflux and influx. Based on comparative transcriptomic analysis of doxorubicin-resistant and -sensitive HCC organoids, the drug-metabolism pathway was found to be related to drug resistance. ABCB1, a well-known drug efflux pump, is upregulated in resistant HCC organoids. ABCB1 inhibitors (e.g., elacridar, encequidar, laniquidar, tariquidar, and valspodar) could improve the sensitivity of doxorubicin, which is not caused by pharmacological toxicity (80) (Fig.2E). In addition, in docetaxel-resistant prostate cancer organoids, PAF1 knockdown significantly reduced overall organoid growth and the number of drug-effluxing (side population) cells (30,81).
ADC drugs targeting tumor cells by exploiting their high expression of membrane proteins have attracted considerable interest from researchers (82,83). However, dynamic changes in tumor cell membrane proteins often cause nonresponsiveness or even resistance to targeted therapy (84, 85). Possible resistance mechanisms include downregulation of membrane proteins, altered internalization, lysosomal degradation, and changed trafficking of ADCs (86). Chen et al. identified resistance mechanisms of anti-HER2 ADCs using transcriptomic data, PDOs, and the I-SPY2 trial (87). SNX10 was significantly downregulated and was closely linked to abnormal intracellular vesicle transport in the resistant PDOs group. Low levels of SNX10 reduced HER2 recycling, increased HER2 trafficking into lysosomes, and decreased cell-surface HER2, leading to resistance to anti-HER2 ADCs (88) (Fig.2E).
Together, these studies provide strong evidence that tumor organoids are an optimal platform for investigating drug resistance (89,90). Elucidating resistance mechanisms through organoid models will aid in the development of predictive biomarkers and improve patient stratification for therapy.
3. Organoid-Guided Exploration of Novel Strategies to Overcome Drug Resistance
Developing new strategies to overcome drug resistance depends on identifying novel therapeutic targets, thereby enabling more effective targeted treatments and ultimately improving patient survival. In addition, combining novel agents with established therapies to create new treatment regimens offers considerable promise for enhancing the sensitivity of conventional treatments. Moreover, the concept of drug repurposing remains highly innovative; agents originally developed for other diseases, when applied outside their traditional contexts, may demonstrate unexpected efficacy against treatment-resistant cancers. Organoids, serving as "drug-testing surrogates," provide a platform for developing innovative therapeutic strategies and offer new insights into reversing tumor resistance (Fig. 3).
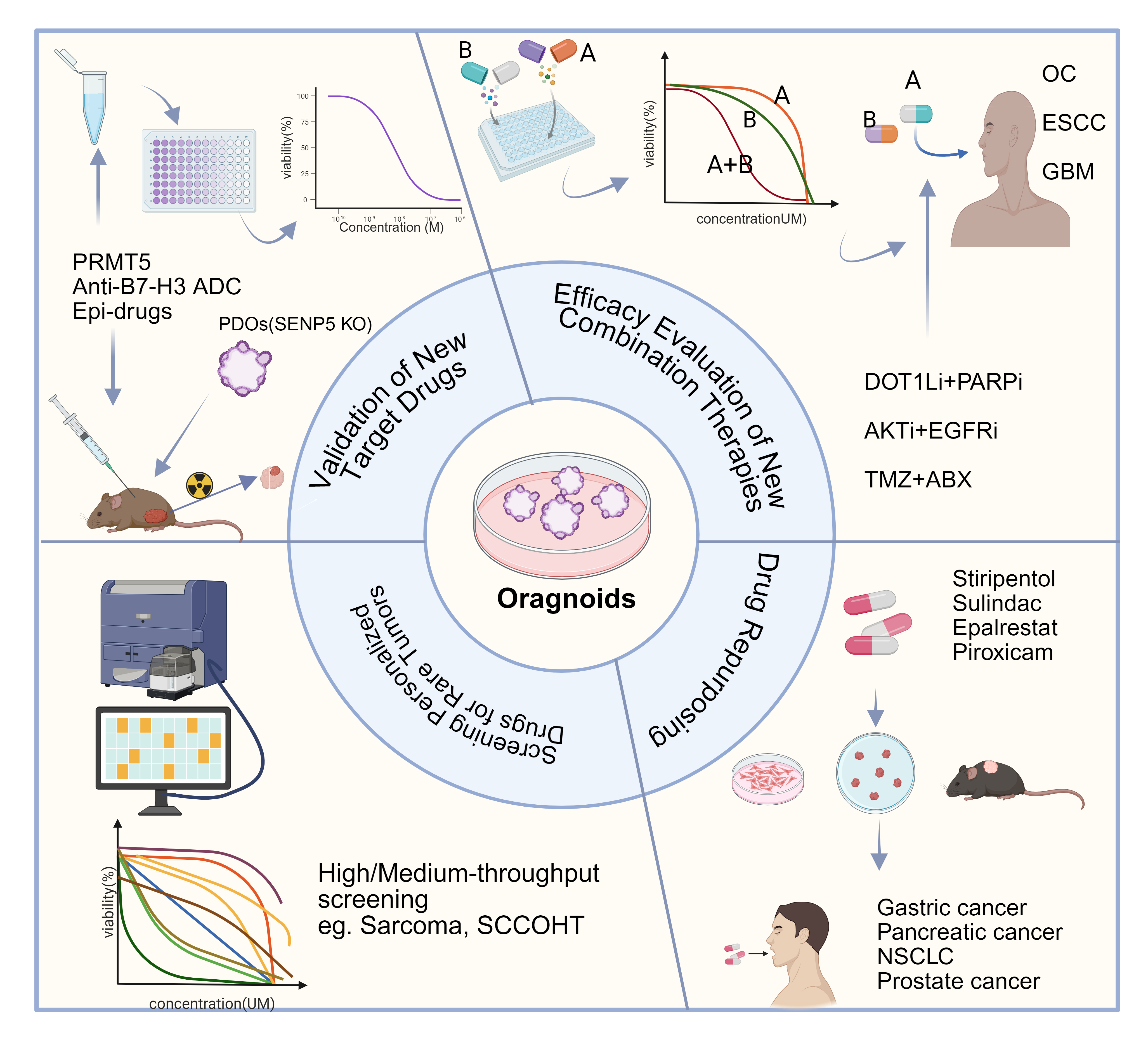
Figure 3. Organoids Guide the Development of New Strategies to Reverse Drug Resistance. Organoids bridge the gap between target discovery in basic research and the clinical application of targeted therapies, serving as a reliable model for validating novel targeted drugs (PRMT5 inhibitor, SENP5 inhibitor, Anti-B7-H3 ADC, Epi-drugs) and new combination treatment regimens. Based on organoid models, the concept of "drug repurposing" can be effectively validated for counteracting drug resistance. Furthermore, high-throughput drug screening using organoids enables the identification of personalized treatment strategies for rare tumors (OC, ovarian cancer). ESCC, esophageal squamous carcinoma. GBM, glioblastoma multiforme. NSCLC, non-small cell lung cancer. SCCOHT, small cell carcinoma of the ovary, hypercalcemic type. KO, Knockdown. TMZ, temozolomide. ABX, albumin-bound paclitaxel. Figure created using BioRender.
3.1 Validation of New Target Drugs
As a potent, radioresistant gene in CRC, SENP5 knockdown, combined with irradiation, significantly inhibited PDO growth, suggesting that SENP5 may be a target to improve radiotherapy efficacy (91). Similarly, cholangiocarcinoma-derived organoids were used to further examine the antitumor effect of the PRMT5 inhibitor. The reduced growth of organoids and the presence of smaller organoids filled with dying cells indicated that PRMT5 may be a clinical therapeutic target (92). With the rapid expansion of antibody–drug conjugate (ADC) therapeutics, organoids offer an ideal platform for evaluating their efficacy (93–95). For example, DS-7300a, an anti-B7-H3 ADC carrying a topoisomerase-1 inhibitor payload, showed strong activity in prostate cancer PDOs harboring specific genetic alterations (96). Moreover, epigenetic drugs (epi-drugs) are small molecules that target epigenetic modifications by modulating enzymes involved in transcriptional and post-transcriptional processes, offering a promising strategy to reverse drug resistance (97,98). In addition, targeting components of the TME represents another emerging approach for developing novel therapies to overcome tumor drug resistance (99).
3.2 Efficacy Evaluation of New Combination Therapies
DOT1L, an H3K79me1/2/3 methylase, was identified as a target of PARPi resistance in OC. SGC0946 (DOT1L inhibitor) could enhance the effect of PARPi (Olaparib and Niraparib) in OC PDOs, with the SGC0946-Olaparib combination showing strong synergistic activity even at low concentrations. Mechanistically, DOT1L inhibition abrogated PLCG2 and ABCB1 upregulation, thereby decreasing PARPi efflux (100). In addition, SIC-19, a novel SIK2 inhibitor, demonstrates synthetic lethality with PARP inhibitors in ovarian cancer PDOs by disrupting RAD50-mediated HR DNA repair (101).
The AKT/mTOR inhibitors were identified as a target hub. In gefitinib-resistant esophageal squamous carcinoma PDOs, an AKT inhibitor demonstrated strong synergy when combined with an EGFR inhibitor (102). Likewise, Qu et al. provided a new perspective on TMZ and albumin-bound paclitaxel (ABX) to augment TMZ sensitivity in GBM. GBM PDOs validated the superior combinatorial efficacy (74). To advance drug discovery aimed at overcoming tumor immunosuppression, T-cell-incorporated pancreatic cancer (PC) organoids have been developed. In this model, T cells are embedded in the outer layer of PC organoids, recreating a physical barrier and enabling detailed analysis of T-cell infiltration and cytotoxicity. Using this system, combinatorial treatment with epigenetic inhibitors and anti-PD-1 demonstrated markedly enhanced anti-tumor effects by upregulating MHC-I antigen processing and presentation and boosting effector T-cell activity (103). Notably, increased drug toxicity remains a major concern when developing multi-agent treatment strategies. Careful optimization of combination regimens, particularly by using reduced or fractionated doses, may help maintain therapeutic efficacy while minimizing adverse effects. Such dose-adjusted combinations also have the potential to lower treatment costs and offer more accessible, cost-effective therapeutic options for patients.
3.3 Drug Repurposing
Another novel approach to explore the drug-sensitization strategy is to repurpose existing compounds (104). Stiripentol has been used clinically as an anti-epileptic treatment (105, 106). As an LDH enzyme inhibitor, it was shown to sensitize PDOs to cisplatin, a finding further confirmed in chemo-resistant gastric cancer patient-derived xenografts (PDX) (75). Additionally, PDOs and PDO-derived xenografts resistant to the nab-paclitaxel plus gemcitabine (AG) regimen were used to demonstrate that sulindac K-80003, a classic anti-inflammatory agent and PI3K/Akt pathway inhibitor, can restore sensitivity to AG therapy in AG-resistant PC (107). Epalrestat, an orally available AKR1B10 inhibitor in clinical use for diabetic polyneuropathy, was successfully repurposed to overcome chemoresistance of non-small cell lung cancer (NSCLC) PDOs (108). In prostate cancer–derived organoid models, piroxicam enhanced the antitumor effects of docetaxel and enzalutamide. Importantly, incorporating PDO-based preclinical testing can strengthen the reliability and translational potential of such combination strategies (109).
These repurposed drugs hold strong potential for advancement into clinical trials for patients with drug-resistant cancers, owing to their well-characterized safety profiles, favorable pharmacokinetics, and established bioavailability. Notably, Chen et al. have already filed a patent application for the use of stiripentol in cancer therapy and have initiated clinical evaluation. A single-arm, prospective phase II trial (ChicTR2400083649) is currently recruiting patients, marking an important step toward translating this repurposed agent into a viable treatment option for resistant malignancies.
3.4 Personalized Drugs Screening for Rare Tumors
For certain rare malignancies, organoids serve as useful in vitro 3D models to investigate optimal drug selection and combination strategies and to recapitulate patient outcomes (110). Shihabi et al. used PDOs to characterize the landscape of drug resistance and sensitivity in sarcoma and developed a high-throughput organoid screening pipeline to identify an FDA-approved or NCCN-recommended effective regimen (111). Patient-derived organoids (PDOs) were successfully generated from small cell carcinoma of the ovary, hypercalcemic type (SCCOHT), enabling high-throughput drug screening. Using a library of 153 clinically approved compounds, organoid-based drug profiling identified methotrexate as a potent and selective agent against SCCOHT, acting by activating the TP53 pathway and inducing apoptosis (112).
These studies underscore that the organoid platform can support the design of therapeutic strategies for drug-resistant and rare cancers.
4. Application of Organoids in Clinical Trials for Drug Resistance
By recapitulating tumor heterogeneity and partially mimicking the tumor microenvironment, organoids provide a powerful platform for advancing drug development (113). Insights derived from basic and preclinical organoid research provide a strong foundation for evaluating targeted inhibitors and novel combination therapies in clinical trials, particularly with respect to therapeutic efficacy and toxicity. As such, the PDO model serves as a promising in vitro system for bridging the gap between preclinical studies and clinical application (114). We summarized the role of organoids in clinical trials by reviewing literature and searching clinical trials involving organoids in ClinicalTrials.gov (Table 1) (Fig.4). Organoids not only select personalized treatment regimens to guide clinical treatment through drug sensitivity tests but also provide new models for exploring tumor drug resistance mechanisms. When integrated into cancer research and clinical decision-making, organoid-based systems represent a major advance toward developing more precise and effective therapies for solid tumors. Their use accelerates the translation of new drugs and therapeutic strategies from the laboratory to the clinic.
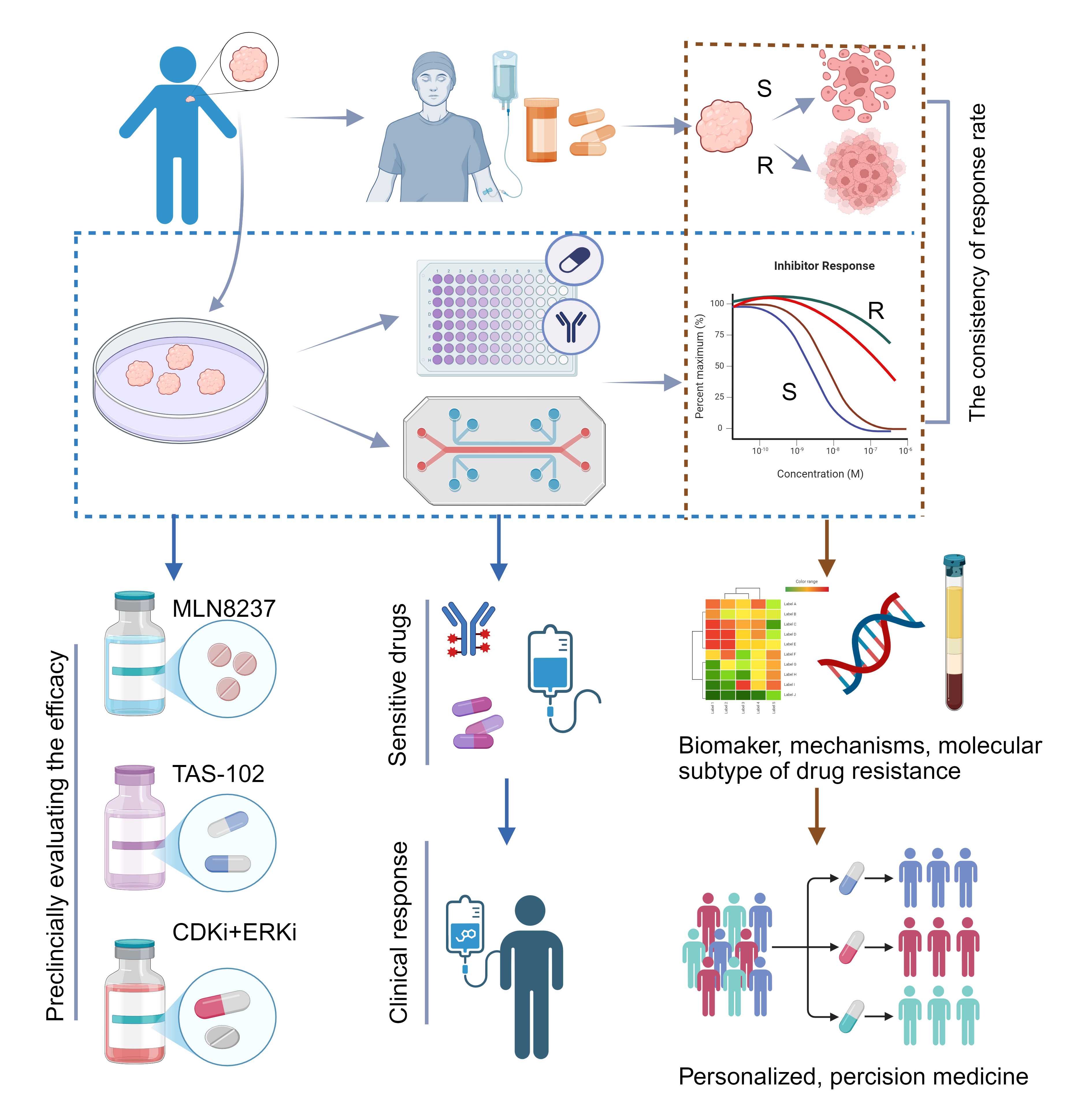
Figure 4. Patient-derived Organoids participate in Clinical Trials. Numerous clinical trials compare the consistency of drug response between organoid-based drug screening and clinical treatment to evaluate the reliability of this platform. Organoid-based drug screening can provide valuable references for clinical decision-making. Organoids can be used as a preclinical model to validate potential targets identified in basic research. By conducting multi-omics analyses on drug-resistant and drug-sensitive organoids, alone or in combination with original tumor tissues, molecular targets and mechanisms underlying resistance can be identified. This approach helps explore molecular subtypes of tumor drug resistance, ultimately guiding precise clinical treatment strategies (R, resistant; S, sensitive). MLN8237, Aurora A kinase inhibitor. TAS-102, nucleoside antitumor agents. Figure created using BioRender.
Table 1. The Application of Patient-derived Organoids (PDOs) in Clinical Trials for Drug Resistance.
Table 1A: Validating the predictive value of PDOs.
| NCT number | Cancer Type | Study type | Country | Start year | Objectives |
|---|---|---|---|---|---|
| NCT05669586 | Lung |
Interventional, Phase 2 |
China | 2023 | Predict therapeutic response in patients with multi-line drug-resistant lung cancer by PDOs |
| NCT04996355 | Colorectal |
Prospective, Observational |
China | 2021 | Explore the feasibility and accuracy of anticancer treatment on organoids-on-a-chip |
| NCT04906733 | Colon |
Prospective, Observational |
China | 2021 | Evaluate the consistency and accuracy of chemotherapy on PDOs to predict the clinical efficacy |
| NCT03979170 | Lung |
Prospective, Observational |
Switzerland | 2019 | Evaluate the consistency and accuracy of chemotherapy on PDOs to predict the clinical efficacy |
| NCT03890614 | Hematologic |
Prospective, Observational |
United States | 2019 | Validate the predictive value of the PDO chemobiogram results compared with retrospective treatment data |
| NCT04859166 | Lung |
Prospective, Observational |
Netherland | 2017 | Prospectively predict treatment response based on primary human lung cancer organoids |
| NCT03251612 |
Metastatic colorectal |
Interventional, Phase 2 |
Denmark | 2017 | Investigate the predictive value of PDOs drug sensitivity testing |
Table 1B: Guiding clinical treatments through PDOs
| NCT number | Cancer Type | Study type | Country | Start year | Objectives |
|---|---|---|---|---|---|
| NCT06739395 | Refractory solid tumors |
Interventional, Phase 2 |
China | 2024 | Organoid as a functional model to test the efficacy of combination therapy based on PD-1/L1 immune checkpoint inhibitors |
| NCT06406608 |
Non-small cell lung cancer |
Interventional | China | 2024 | PDOs drug sensitivity test guided clinical treatment plans for drug-resistant/relapsed conditions |
| NCT06315868 | Breast cancer |
Prospective, Observational |
Italy | 2023 | Exploring the molecular phenotype associated with chemoresistance through PDOs |
| NCT05532397 | High grade astrocytic glioma | Interventional | Singapore | 2023 | Screening optimal drug combinations for patients with recurrent conditions through PDOs |
| NCT05813509 | Ovarian cancer | Interventional | China | 2022 | Screening potential clinical therapeutic drugs for patients with resistant ovarian cancer through PDOs |
Table 1C: Exploring drug resistance mechanisms
| NCT number | Cancer Type | Study type | Country | Start year | Objectives |
|---|---|---|---|---|---|
| NCT06791941 | Head and Neck cancer | Retrospective, Non-intervention | Italy | 2024 | Characterizing molecular networks involved in resistance and develop new effective therapeutic strategies based on organoids |
| NCT06155370 | Gynecological tumors |
Cross-Sectional Observational |
China | 2023 | Construction of organoids biobank for exploring the molecular mechanism of tumor pathogenesis and resistance |
| NCT06156150 | Glioma | Observational | China | 2023 | Use of organoids to confirm the mechanism of treatment resistance of vaccines |
| NCT05518110 | Refractory pancreatic cancer |
Interventional, Phase 2 |
Ireland | 2023 | Exploring primary and emerging resistance mechanisms in patients with metastatic refractory pancreatic cancer treated with trametinib and hydroxychloroquine |
| NCT04504747 | Breast cancer | Prospective, Observational | France | 2022 | Identifying robust candidates for drug resistance upon PDOs mimicking patient's response |
| NCT04868396 | Glioblastoma | Prospective, Observational | Netherland | 2021 | Establishing PDOs to study study mechanisms that contribute to aggressive tumor growth and treatment resistance |
| NCT04526587 | Breast cancer | Prospective, Observational | United States | 2020 | Developing PDOs from resistant disease to functionally assess the mechanisms occurring with resistance. |
| NCT03925233 | Breast cancer |
Retrospective Observational |
China | 2019 | Sensitivity detection and drug resistance mechanism of therapeutic drugs based on PDOs |
| NCT03010722 | Metastatic colorectal cancer | Prospective, Observational | United Kingdom | 2015 | Investigating molecular predictors of resistance and response to regorafenib therapy |
| NCT04846933 | Ovarian cancer | Interventional | Finland | 2012 | Multi-layer Data from fresh tissue and PDOs to predict therapy resistance and explore new therapeutic plans |
Table 1D: Preclinically evaluating the efficacy of novel therapy strategies
| NCT number | Cancer Type | Study type | Country | Start year | Objectives |
|---|---|---|---|---|---|
| NCT03454035 | Pancreatic and KRAS-mutant colorectal cancer |
Interventional, Phase 1 |
United States | 2018 | Evaluating the efficacy of the ERK inhibitor (ulixertinib) in combination with the CDK4/6 inhibitor (palbociclib) on PDOs |
| NCT01799278 | Neuroendocrine prostate cancer |
Interventional, Phase 2 |
United States | 2013 | Comparing the response of alisertib (MLN8237) on PDOs and clinical patients |
| NCT01607957 |
Metastatic colorectal cancer |
Interventional, Phase 3 |
United States | 2012 | Validating the association of KRASG12 and intrifluridine resistance |
4.1 Validating the predictive value of PDOs
A total of 25 clinical trials involving organoids were reviewed, seven of which aim to predict clinical therapeutic responses using 3D organoids. The trials NCT03890614, NCT03979170, NCT05669586, and NCT04859166 validate the predictive value of 3D organoid chemobiograms compared with retrospective and prospective data on donors’ therapeutic responses. Sensitivity to cetuximab and its correlation with PDOs and clinical responses were evaluated
to predict treatment options for patients with RAS/RAF wild-type CRC (NCT04906733). Specifically, trial NCT04996355 was designed to validate the predictive ability of organoid-on-chip systems for drug screening. PDOs are believed to aid in selecting personalized treatment plans for patients with multilined, drug-resistant conditions. Jensen et al. conducted a clinical trial (NCT03251612) to assess the predictive value of drug testing in CRC PDOs, to identify effective therapeutic regimens based on CRC organoids. Results showed that 2-month progression-free survival (PFS) was achieved in 17 patients (50%; 95% CI: 32-68), exceeding the predefined threshold (14 of 45; 31%) (115).
These studies demonstrate that organoids hold promise for advancing personalized medicine and guiding treatment in newly diagnosed and relapsed-resistant malignancies. However, they are limited by the absence of the tumor microenvironment (TME) and immune cells (116). The development of specialized assays to examine interactions between cancer cells and components of the microenvironment may improve the accuracy of in vitro testing.
4.2 Guiding clinical treatments through PDOs
Three ongoing interventional studies (NCT05813509, NCT06406608, and NCT05532397) are using organoid-based drug-sensitivity screening to identify effective therapies for resistant or recurrent ovarian cancer, non–small cell lung cancer, and high-grade astrocytic glioma, respectively. In breast cancer, efforts to integrate drug-sensitivity data with molecular subtypes of resistance may further refine treatment selection (NCT06315868).
The combined use of genomics-driven precision medicine and organoid-based functional testing is emerging as a feasible strategy for guiding certain precision chemotherapy approaches (117). For example, Wang et al. launched a precision-medicine trial (NCT06739395) employing molecularly matched therapies for patients who had exhausted standard treatment options.
In clinical practice, molecular tumor boards, which rely on sequencing data, clinical judgment, and published literature, are not always able to match patients to targeted therapies for their specific genomic alterations. In such situations, functional models, such as organoids, can provide an additional layer of decision-making, offering patients a personalized opportunity to evaluate potential therapeutic interventions when no molecular match is available.
4.3 Exploring drug resistance mechanisms
Organoid biobank–guided drug-sensitivity profiling not only provides a platform for tailoring clinical treatments but also enables systematic investigation of drug-resistance mechanisms (NCT06155370). For example, breast cancer PDOs generated from patients with CDK4/6 inhibitor-resistant disease are being used to dissect mechanisms of resistance (NCT04526587) functionally.
Chen et al. initiated a clinical study (NCT06156150) to investigate the functional role and resistance mechanisms associated with the B7-H4/ATF3 axis in glioma. An organoid platform was employed to validate the role of macrophage-derived B7-H4 in influencing chemokine secretion for T cells and in contributing to resistance to vaccine-based therapies (NCT04868396).
The PaTcH study (NCT05518110) aims to characterize both primary and acquired resistance mechanisms in patients with metastatic, treatment-refractory pancreatic cancer undergoing combined trametinib and hydroxychloroquine therapy. PC organoids are established for each patient before and during treatment, providing ex vivo models to trace the evolution of resistance and to screen potential therapeutic agents.
Together, these studies highlight the expanding clinical integration of organoid platforms for both personalized therapeutic guidance and mechanistic exploration of drug resistance.
In addition, combining molecular analyses (tissue and PDOs) with therapeutic response (clinical response and PDOs-guided drug testing) may facilitate understanding of drug resistance mechanisms. In the PROSPECT-R trial (NCT03010722), large-scale microRNA expression analysis of matched liquid and tissue biopsies found MIR652-3p as a biomarker of regorafenib response. Tissue biopsies collected at baseline, after 2 months, and at progression were used to establish PDOs and perform molecular analyses. Moreover, PDO co-cultures and PDO-xenotransplants were generated for functional analyses, which showed that MIR652-3p controlled resistance to regorafenib by impairing regorafenib-induced autophagy and orchestrating a switch from neo-angiogenesis to vessel co-option (118).
The study (NCT06791941) aims to characterize the molecular networks underlying resistance in head and neck tumors driven by mutated p53/YAP, and to develop new therapeutic strategies to target these networks and enhance the effectiveness of current treatments.
The DECIDER project (NCT04846933) will develop AI-powered diagnostic tools, advanced software platforms for clinical decision-making, new data analysis and integration techniques, and high-throughput drug-screening methods in organoids. The multi-layer data could uncover key mechanisms underlying chemoresistance in HGSOC patients and inform the development of personalized treatment strategies for chemotherapy-resistant patients.
4.4 Preclinically evaluating the efficacy of novel therapy strategies
PC PDOs treated with alisertib (MLN8237) show concordant responses with their corresponding patients (NCT01799278). In the RECOURSE trial (NCT01607957), whole-genome analysis identified KRAS codon G12 mutations as a potential biomarker of therapeutic resistance. Consistent with these findings, CRC PDO studies showed that KRAS G12 mutations were associated with an increased resistance to trifluridine-based genotoxic therapies. (119, 120).
Organoids capable of dynamically measuring Aurora–N-myc complex activity provide a powerful platform for identifying patients most likely to benefit from single-agent alisertib. By evaluating pathway dependency and drug responsiveness in real time, organoid-based functional testing can optimize patient selection, enhance treatment precision, and increase the likelihood of clinical benefit from Aurora kinase–targeted strategies (121).
Organoid models have also been key in identifying effective combination therapies. For example, CDK4/6 and ERK inhibitors cooperatively reduced proliferation and induced apoptosis in both PC organoids and KRAS-mutant CRC organoids. This combination suppressed CDK4/6 inhibition–induced compensatory activation of ERK, PI3K, anti-apoptotic signaling, and MYC expression. These preclinical findings supported a Phase I clinical trial (NCT03454035) evaluating the ERK inhibitor ulixertinib in combination with the CDK4/6 inhibitor palbociclib in patients with advanced pancreatic cancer.
Together, these studies illustrate how organoid validation provides essential guidance for clinical translation. The organoid platform can be leveraged to test novel combinations of small-molecule inhibitors with standard therapies, accelerating the development of precision treatments for drug-resistant malignancies (122). The use of organoids in clinical research remains in the early stages. Organoid-guided drug sensitivity testing helps identify potential therapies for recurrent or resistant tumors and supports the development of new drugs. Additionally, organoids can generate multi-omics data that aid in establishing molecular subtypes of drug resistance and in discovering and validating biomarkers for resistance.
Discussion
Tumor drug resistance is a primary cause of failure in cancer treatment. Traditional models for studying tumor drug resistance, such as cell lines and animal experiments, have significant limitations. Conversely, organoids can closely replicate tissue architecture and cellular diversity, providing unique advantages in studying drug resistance (123-125). As a preclinical model, organoids with high fidelity serve as a novel platform for examining drug resistance. Recent progress includes investigating resistance mechanisms in basic research, evaluating new treatment regimens in preclinical models, and participating in clinical trials to accelerate the translation of new drugs or therapies into clinical practice.
Because of the absence of microenvironment components, an organoid-based coculture system involving immune cells, non-immune cells, and tumor cells was developed to explore the role of the TME in drug resistance. Co-culturing organoids with stromal cells to mimic the in vivo immune microenvironment of primary tumors can more accurately reflect drug responses. Recently, vascularization and immune microenvironment modeling using organoid technology have become a focus of research. TME (such as immune cells, fibroblasts, and vascular networks) plays a key role in drug resistance. Cocultures of gastric cancer organoids and immune cells were used to study the immunosuppressive function of myeloid-derived suppressor cells (MDSCs), showing that PD-L1 expression is controlled by the mTOR signaling pathway in gastric cancer (126). Additionally, increased tumor growth and reduced cytotoxic T-cell proliferation were observed in cocultures of PCOs with MDSCs (127). Overall, improving organoid technology through coculture systems could be a promising in vitro platform for modeling the tumor immune environment.
Currently, organoids are being used to investigate and validate mechanisms of drug resistance (128,129). Additionally, organoids could serve as an excellent platform for testing novel and promising combination treatment strategies to overcome drug resistance. Here, seven hepatobiliary tumor organoids were generated to investigate heterogeneity and evolution using single-cell RNA sequencing. HCC272, which exhibits a high level of epithelial-mesenchymal transition, shows broad-spectrum drug resistance (130). Given advances in sequencing and multi-omics analysis approaches combined with patient-derived iPSC models and gene-editing platforms, substantial progress has been made in diagnosing and treating rare and undiagnosed diseases. These techniques provide a solid foundation for future precision medicine studies (131). Although organoids have entered clinical trials, their role remains primarily in basic validation and early exploration of drug resistance mechanisms. Due to ethical constraints surrounding organoids, there are currently few interventional studies using organoids to guide clinical treatment for patients with rare tumors or those facing multidrug-resistant relapses. Treatment strategies based on organoid drug screening can guide clinical therapy, provided patients give full informed consent.
Limitations and critiques of PDO models
Although organoids are hailed as "near-physiological organ analogs" and demonstrate significant potential in fundamental cancer research and clinical applications, several critical bottlenecks and challenges remain. Primarily, the establishment, maintenance, and passage of organoids incur substantial costs and lack standardized, optimized methods. Current success rates in generating organoids for various cancer types show considerable variability (132-134). This can lead to poor reproducibility of organoid-based applications. Additionally, the absence of environmental components and a vascular network may hinder accurate modeling and prediction of therapeutic responses and limit their widespread use in studies of drug resistance (135-137). Thirdly, the original tumor tissue used to derive organoids reflects only local genetic characteristics, resulting in differences between in vivo and in vitro conditions. Therefore, sample heterogeneity adds complexity and hampers validation of the predictive value of clinical efficacy in organoids. Sampling tissue from multiple regions of the same tumor may better capture tumor heterogeneity and more reliably advance translational cancer research. Due to limitations of organoids and the restricted application of emerging experimental techniques in organoid models, their use in studying tumor drug resistance and in clinical trials remains straightforward. In-depth research on drug resistance will likely require breakthroughs in organoid and experimental technologies.
Future directions and technological integration
An increasing number of innovative technologies are emerging to overcome these limitations and leverage the inherent strengths of organoids, thereby enabling in-depth research into tumor drug resistance using organoid models. The recent revolution in genome-editing technologies (CRISPR-Cas9) offers a platform to explore the effects of ‘repairing’ disease-causing genes. Organoids can be used to test whether repair of a specific oncogenic mutation reverses the tumorigenic phenotype (138,139). The CRISPR-Cas9 strategy has been employed to generate mutant organoid lines or to correct the gene locus via homologous recombination in organoids. These edited organoids provide a valuable tool for studying mechanisms of drug resistance (140,141). A platform for pooled CRISPR-Cas9 screening in human colon organoids proves useful for patient-specific functional genomics (142).
Furthermore, new sequencing techniques could be applied to organoid models, including single-cell sequencing, multi-omics analysis, and spatial transcriptomics, thereby facilitating in-depth research on drug resistance (143,144). The combination of brain organoids and single-cell sequencing uncovers molecular and cellular abnormalities induced by microgravity during human brain development (145). Innovative technologies based on organoids may offer new insights into the molecular networks underlying resistance and into the development of targeted drugs for clinical use. Additionally, the integration of organoids and artificial intelligence (AI) is an emerging approach that aims to build more complex and accurate models of human diseases and serve as a powerful tool for drug discovery, disease diagnosis, and treatment development (146). AI-assisted organoid drug screening allows high-throughput, automated, and standardized processes, which are expected to improve the efficiency of preclinical screening and accelerate clinical translation. Organ-on-a-chip (OoC) technology, which faithfully reproduces organ-level physiology and pathophysiology, holds significant promise for advancing drug development (147-150). We anticipate that coupling OoC and AI will further reveal mechanisms of drug resistance and enhance drug discovery (146, 151).
Additionally, because organoids lack specific essential TME components, 3D bioprinters have emerged as tools to create high-resolution microstructures that mimic TME complexity, thereby offering a precise model for personalized anti-cancer drug screening (152,153). It has been reported that 3D bioprinting technology was used to accurately produce kidney and breast organoids for drug assessment and screening (154,155). Moreover, co-culture systems and microfluidic chip systems based on organoids have become key areas of focus for developing biomimetic microenvironments and diverse cell populations (156-159). Additionally, angiogenesis, a vital component of the TME, plays a pivotal role in tumor growth and resistance by supplying nutrients and facilitating tumor cell dissemination (160, 161). Alongside an improved co-culture system, a microfluidic platform integrating functional vascularized organoids-on-a-chip has been used to establish organoid perfusion via advanced microfluidics (162). Current efforts to vascularize organoids still face challenges in accurately mimicking tumor vasculature, hindering studies of tumor progression and therapeutic response (163).
With the emergence of the novel technologies mentioned above, organoids are increasingly important in regenerative medicine. These emerging uses of organoids create new opportunities for rapid multiplex drug screening and developing personalized cancer models, signaling a major shift in drug resistance studies and pharmaceutical progress. This will encourage broader and more thorough use of organoids in clinical trials, thereby shortening development timelines and reducing costs. As understanding of tumor drug resistance mechanisms advances, molecular subtypes associated with resistance across various cancers will become more precise. Molecular subtype-guided clinical diagnosis and treatments will further enhance precision medicine practices.
References
1. Wise JF, Lawrence MS. Huge whole-genome study of human metastatic cancers. Nature 2019;575:60–61. https://doi.org/10.1038/d41586-019-03123-0
2. Xing P, Wang S, Cao Y, Liu B, Zheng F, Guo W, et al. Treatment strategies and drug resistance mechanisms in adenocarcinoma of different organs. Drug Resist Updat. 2023;71:101002. https://doi.org/10.1016/j.drup.2023.101002
3. Nussinov R, Tsai CJ, Jang H. Anticancer drug resistance: an update and perspective. Drug Resist Updat. 2021;59:100796. https://doi.org/10.1016/j.drup.2021.100796
4. Peng Z, Lv X, Sun H, Zhao L, Huang S. 3D tumor cultures for drug resistance and screening development in clinical applications. Mol Cancer. 2025;24:93. https://doi.org/10.1186/s12943-025-02281-2
5. Chai C, Ji P, Xu H, Tang H, Wang Z, Zhang H, et al. Targeting cancer drug resistance utilizing organoid technology. Biomed Pharmacother. 2023;158:114098. https://doi.org/10.1016/j.biopha.2022.114098
6. Rahmanian M, Seyfoori A, Ghasemi M, Shamsi M, Rezaei Kolahchi A, Pezeshgi Modarres H, et al. In-vitro tumor microenvironment models containing physical and biological barriers for modelling multidrug resistance mechanisms and multidrug delivery strategies. J Control Release. 2021;334:164–177. https://doi.org/10.1016/j.jconrel.2021.04.024
7. Wang Q, Yuan F, Zuo X, Li M. Breakthroughs and challenges of organoid models for assessing cancer immunotherapy: a cutting-edge tool for advancing personalised treatments. Cell Death Discov. 2025;11:222. https://doi.org/10.1038/s41420-025-02505-w
8. Singh D, Thakur A, Rakesh, Kumar A. Advancements in organoid-based drug discovery: revolutionizing precision medicine and Pharmacology. Drug Dev Res. 2025;86:e70121. https://doi.org/10.1002/ddr.70121
9. Wang Y, Sun X, Lu B, Zhang D, Yin Y, Liu S, et al. Current applications, future Perspectives and challenges of Organoid technology in oral cancer research. Eur J Pharmacol. 2025;993:177368. https://doi.org/10.1016/j.ejphar.2025.177368
10. Li Z, Li K, Zhang C, Zhao Y, Guo Y, He J, et al. Bioprinted organoids: an innovative engine in biomedicine. Adv Sci (Weinh). 2025;12(33):e07317. https://doi.org/10.1002/advs.202507317
11. El Harane S, Nazari B, El Harane N, Locatelli M, Zidi B, Durual S, et al. Generation of individualized, standardized, and electrically synchronized human midbrain organoids. Cells. 2025;14(15):1211. https://doi.org/10.3390/cells14151211
12. Wu J, Liu T, Zhang X, Qu C, Wu J, Xu S, et al. Progress in the application of organoids for exploring the relationship between macrophages and various lung diseases. Biofabrication. 2025;17(3). https://doi.org/10.1088/1758-5090/adde15
13. Ding Z, Chang X, Qu X, Hua K, Qiu J. Gynecological malignancy organoids: A game changer for personalized medicine. Biochim Biophys Acta Rev Cancer. 2025;1880:189405. https://doi.org/10.1016/j.bbcan.2025.189405
14. Li Q, Xiao Y, Han L, Luo W, Dai W, Fang H, et al. Microbiome dysbiosis, neutrophil recruitment and mesenchymal transition of mesothelial cells promotes peritoneal metastasis of colorectal cancer. Nat Cancer. 2025;6(3):493-510. https://doi.org/10.1038/s43018-025-00910-9
15. Boilève A, Cartry J, Goudarzi N, Bedja S, Mathieu JRR, Bani MA, et al. Organoids for functional precision medicine in advanced pancreatic cancer. Gastroenterology. 2024;167(5):961-976.e13. https://doi.org/10.1053/j.gastro.2024.05.032
16. Sun X, Cai W, Li H, Gao C, Ma X, Guo Y, et al. Endothelial-like cancer-associated fibroblasts facilitate pancreatic cancer metastasis via vasculogenic mimicry and paracrine signalling. Gut. 2025;74(9):1437-1451. https://doi.org/10.1136/gutjnl-2024-333638
17. Yang R, Wang S, Li Z, Yin C, Huang W, Huang W. Patient-derived organoid co-culture systems as next-generation models for bladder cancer stem cell research. Cancer Lett. 2025;625:217793. https://doi.org/10.1016/j.canlet.2025.217793
18. Li Y, Liu J, Xu S, Wang J. 3D bioprinting: an important tool for tumor microenvironment research. Int J Nanomedicine. 2023;18:8039-8057. https://doi.org/10.2147/IJN.S435845
19. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19(2):65-81. https://doi.org/10.1038/s41568-018-0104-6
20. Mi W, van Tienderen GS, Shi S, Broeders A, Monfils K, Roest HP, et al. Apoptosis regulators of the Bcl-2 family play a key role in chemoresistance of cholangiocarcinoma organoids. Int J Cancer. 2025;157(8):1694-1708. https://doi.org/10.1002/ijc.35483
21. Álvarez-Varela A, Novellasdemunt L, Barriga FM, Hernando-Momblona X, Cañellas-Socias A, Cano-Crespo S, et al. Mex3a marks drug-tolerant persister colorectal cancer cells that mediate relapse after chemotherapy. Nat Cancer. 2022;3(9):1052-1070. https://doi.org/10.1038/s43018-022-00402-0
22. Yang R, Kwan W, Du Y, Yan R, Zang L, Li C, et al. Drug-induced senescence by aurora kinase inhibitors attenuates innate immune response of macrophages on gastric cancer organoids. Cancer Lett. 2024;598:217106. https://doi.org/10.1016/j.canlet.2024.217106
23. Würth R, Donato E, Michel LL, Saini M, Becker L, Cheytan T, et al. Circulating tumor cell plasticity determines breast cancer therapy resistance via neuregulin 1-HER3 signaling. Nat Cancer. 2025;6(1):67-85. https://doi.org/10.1038/s43018-024-00882-2
24. Tong X, Patel AS, Kim E, Li H, Chen Y, Li S, et al. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. Cancer Cell. 2024;42(3):413-428.e7. https://doi.org/10.1016/j.ccell.2024.01.012
25. Sase M, Sato T, Sato H, Miya F, Zhang S, Haeno H, et al. Comparative analysis of tongue cancer organoids among patients identifies the heritable nature of minimal residual disease. Dev Cell. 2025;60(3):396-413.e6. https://doi.org/10.1016/j.devcel.2024.10.007
26. Mosquera MJ, Kim S, Bareja R, Fang Z, Cai S, Pan H, et al. Extracellular matrix in synthetic hydrogel-based prostate cancer organoids regulate therapeutic response to EZH2 and DRD2 inhibitors. Adv Mater. 2022;34(2):e2100096. https://doi.org/10.1002/adma.202100096
27. Yang D, Zhang X, Hu Z, Sun Q, Fu H, Yao J, et al. Organoid-based single cell sequencing revealed the lineage evolution during docetaxel treatment in gastric cancer. Cancer Lett. 2025;619:217617. https://doi.org/10.1016/j.canlet.2025.217617
28. Mancini C, Lori G, Mattei G, Iozzo M, Desideri D, Cianchi F, et al. PHGDH drives 5-FU chemoresistance in colorectal cancer through the Hedgehog signaling. J Exp Clin Cancer Res. 2025;44(1):198. https://doi.org/10.1186/s13046-025-03447-y
29. Xu Q, Jiang Z, Pan Y, Li S, Cao Z, Hua S, et al. Cucurbitacin B stimulates PD-1 immunotherapy response in malignant breast cancer by covalent targeting MTCH2. 2025;145:157017. https://doi.org/10.1016/j.phymed.2025.157017
30. Muniyan S, Vengoji R, Nimmakayala RK, Seshacharyulu P, Perumalsamy B, Alsafwani ZW, et al. PAF1-mediated transcriptional reprogramming confers docetaxel resistance in advanced prostate cancer. Cancer Lett. 2025;609:217355. https://doi.org/10.1016/j.canlet.2024.217355
31. Fei Y, Cao D, Li Y, Wang Z, Dong R, Zhu M, et al. Circ_0008315 promotes tumorigenesis and cisplatin resistance and acts as a nanotherapeutic target in gastric cancer. J Nanobiotechnology. 2024;22:519. https://doi.org/10.1186/s12951-024-02760-6
32. Ren T, Qiu J, Chen F, Jiang Q, Liu Q, Wu T, et al. Targeting glutamine metabolism transporter SLC25A22 enhances CD8+ T-cell function and anti-PD-1 therapy efficacy in cervical squamous cell carcinoma: integrated metabolomics, transcriptomics and T-cell-incorporated tumor organoid studies. Adv Sci (Weinh). 2025;12:e02225. https://doi.org/10.1002/advs.202502225
33. Yao N, Jing N, Lin J, Niu W, Yan W, Yuan H, et al. Patient-derived tumor organoids for cancer immunotherapy: culture techniques and clinical application. Invest New Drugs. 2025;43:394–404. https://doi.org/10.1007/s10637-025-01523-w
34. Goto H, Nishioka Y, et al. Goto H, Nishioka Y. Fibrocytes: a novel stromal cells to regulate resistance to anti-angiogenic therapy and cancer progression. Int J Mol Sci. 2017;19:98. https://doi.org/10.3390/ijms19010098
35. Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. https://doi.org/10.1038/s41591-018-0096-5
36. Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, et al. Cancer-associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell Mol Gastroenterol Hepatol. 2021;11:407–431. https://doi.org/10.1016/j.jcmgh.2020.09.003
37. Guo Y, Li Q, Ye Q, Jin Y, Yu Y, Zhang X, et al. Construction and drug screening of co-culture system using extrahepatic cholangiocarcinoma organoids and tumor-associated macrophages. Heliyon. 2024;10:e36377. https://doi.org/10.1016/j.heliyon.2024.e36377
38. Kang SH, Oh SY, Lee KY, Lee HJ, Kim MS, Kwon TG, et al. Differential effect of cancer-associated fibroblast-derived extracellular vesicles on cisplatin resistance in oral squamous cell carcinoma via miR-876-3p. Theranostics. 2024;14:460–479. https://doi.org/10.7150/thno.87329
39. Farin HF, Mosa MH, Ndreshkjana B, Grebbin BM, Ritter B, Menche C, et al. Colorectal cancer organoid-stroma biobank allows subtype-specific assessment of individualized therapy responses. Cancer Discov. 2023;13:2192–2211. https://doi.org/10.1158/2159-8290.CD-23-0050
40. Ma Y, Xue F, Pei Z, Zhao Y. Constructing a co-culture model of cancer-associated fibroblasts and ovarian cancer organoids and studying mechanisms of drug resistance. Exp Cell Res. 2025;450:114656. https://doi.org/10.1016/j.yexcr.2025.114656
41. Ma B, Ma C, Long X, Jiang L. Stromal reprogramming in urachal cancer: fibroblast activation protein and collagen remodeling drive immune-suppressive niches and immunotherapy resistance. Int Immunopharmacol. 2025;163:115204. https://doi.org/10.1016/j.intimp.2025.115204
42. Ryu KB, Seo JA, Lee K, Choi J, Yoo G, Ha JH, et al. Drug-resistance biomarkers in patient-derived colorectal cancer organoid and fibroblast co-culture system. Curr Issues Mol Biol. 2024;46:5794–5811. https://doi.org/10.3390/cimb46060346
43. Roh HS, Kim DE, Kim G, Kim J, Fan D, Kim HS, et al. Establishment and long-term expansion of adult hepatobiliary organoids co-cultured with liver endothelial cells. Heliyon. 2024;10:e36120. https://doi.org/10.1016/j.heliyon.2024.e36120
44. Lin CN, Liang YL, Tsai HF, Wu PY, Huang LY, Lin YH, et al. Adipocyte pyroptosis occurs in omental tumor microenvironment and is associated with chemoresistance of ovarian cancer. J Biomed Sci. 2024;31:62. https://doi.org/10.1186/s12929-024-01051-4
45. Jiang S, Deng T, Cheng H, Liu W, Shi D, Yuan J, et al. Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J Exp Clin Cancer Res. 2023;42:199. https://doi.org/10.1186/s13046-023-02756-4
46. Shi F, Li GJ, Liu Y, Zhou HM, Zhang Y, Wei SY, et al. USP19 deficiency enhances T-cell-mediated antitumor immunity by promoting PD-L1 degradation in colorectal cancer. Pharmacol Res. 2025;214:107668. https://doi.org/10.1016/j.phrs.2025.107668
47. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598.e12. https://doi.org/10.1016/j.cell.2018.07.009
48. Subtil B, Cambi A, Tauriello DVF, de Vries IJM. The therapeutic potential of tackling tumor-induced dendritic cell dysfunction in colorectal cancer. Front Immunol. 2021;12:724883. https://doi.org/10.3389/fimmu.2021.724883
49. LeSavage BL, Zhang D, Huerta-López C, Gilchrist AE, Krajina BA, Karlsson K, et al. Engineered matrices reveal stiffness-mediated chemoresistance in patient-derived pancreatic cancer organoids. Nat Mater. 2024;23:1138–1149. https://doi.org/10.1038/s41563-024-01908-x
50. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e16. https://doi.org/10.1016/j.cell.2018.11.021
51. Zhang B, Ohuchida K, Tsutsumi C, Shimada Y, Mochida Y, Oyama K, et al. Dynamic glycolytic reprogramming effects on dendritic cells in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2024;43:271. https://doi.org/10.1186/s13046-024-03192-8
52. Zhang J, Tavakoli H, Ma L, Li X, Han L, Li X. Immunotherapy discovery on tumor organoid-on-a-chip platforms that recapitulate the tumor microenvironment. Adv Drug Deliv Rev. 2022;187:114365. https://doi.org/10.1016/j.addr.2022.114365
53. Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. https://doi.org/10.1158/2159-8290.CD-17-0833
54. Sethakorn N, Heninger E, Breneman MT, Recchia E, Ding AB, Jarrard DF, et al. Integrated analysis of the tumor microenvironment using a reconfigurable microfluidic cell culture platform. FASEB J. 2022;36:e22540. https://doi.org/10.1096/fj.202200684RR
55. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. https://doi.org/10.1038/nbt.1685
56. Gunjan A, Singh RK. Epigenetic therapy: targeting histones and their modifications in human disease. Future Med Chem. 2010;2:543–548. https://doi.org/10.4155/fmc.10.18
57. Orsolic I, Carrier A, Esteller M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet. 2023;39:74–88. https://doi.org/10.1016/j.tig.2022.10.004
58. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. https://doi.org/10.1016/j.cell.2017.05.045
59. Wang D, Zhang Y, Li Q, Zhang A, Xu J, Li Y, et al. N6‑methyladenosine (m6A) in cancer therapeutic resistance: potential mechanisms and clinical implications. Biomed Pharmacother. 2023;167:115477. https://doi.org/10.1016/j.biopha.2023.115477
60. Kraiczy J, Nayak KM, Howell KJ, Ross A, Forbester J, Salvestrini C, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. https://doi.org/10.1136/gutjnl-2017-314817
61. Ghorbaninejad M, Asadzadeh‑Aghdaei H, Baharvand H, Meyfour A. Intestinal organoids: a versatile platform for modeling gastrointestinal diseases and monitoring epigenetic alterations. Life Sci. 2023;319:121506. https://doi.org/10.1016/j.lfs.2023.121506
62. Nguyen TBN, Gevers S, Kok RNU, Burgering LM, Neikes H, Akkerman N, et al. Lactate controls cancer stemness and plasticity through epigenetic regulation. Cell Metab. 2025;37:903–919.e10. https://doi.org/10.1016/j.cmet.2025.01.002
63. Silva‑Almeida C, Ewart MA, Wilde C. 3D gastrointestinal models and organoids to study metabolism in human colon cancer. Semin Cell Dev Biol. 2020;98:98–104. https://doi.org/10.1016/j.semcdb.2019.05.019
64. Yu M, Ni M, Xu F, Liu C, Chen L, Li J, et al. NSUN6‑mediated 5‑methylcytosine modification of NDRG1 mRNA promotes radioresistance in cervical cancer. Mol Cancer. 2024;23:139. https://doi.org/10.1186/s12943-024-02055-2
65. Zhang X, Su T, Wu Y, Cai Y, Wang L, Liang C, et al. N6‑methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. 2024;84:827–840. https://doi.org/10.1158/0008-5472.CAN-23-1916
66. Xie R, Cheng L, Huang M, Huang L, Chen Z, Zhang Q, et al. NAT10 drives cisplatin chemoresistance by enhancing ac4C‑associated DNA repair in bladder cancer. Cancer Res. 2023;83:1666–1683. https://doi.org/10.1158/0008-5472.CAN-22-2233
67. Xu X, Wang Q, Guo K, Xu J, Lu Y, Chen H, et al. CD47 blockade reverses resistance to HDAC inhibitor by liberating anti‑tumor capacity of macrophages. J Exp Clin Cancer Res. 2025;44:67. https://doi.org/10.1186/s13046-025-03335-5
68. Zhang M, Zhong A, Liu H, Zhao L, Wang Y, Lu Z, et al. EZH2 loss promotes gastric squamous cell carcinoma. Nat Commun. 2025;16:6032. https://doi.org/10.1038/s41467-025-61024-5
69. Wang L, Wang X, Zhu X, Zhong L, Jiang Q, Wang Y, et al. Drug resistance in ovarian cancer: from mechanism to clinical trial. Mol Cancer. 2024;23:66. https://doi.org/10.1186/s12943-024-01967-3
70. Ikeuchi H, Matsuno Y, Kusumoto‑Matsuo R, Kojima S, Ueno T, Ikegami M, et al. GLI1 confers resistance to PARP inhibitors by activating the DNA damage repair pathway. Oncogene. 2024;43:3037–3048. https://doi.org/10.1038/s41388-024-03105-1
71. Ubhi T, Zaslaver O, Quaile AT, Plenker D, Cao P, Pham NA, Békési A, et al. Cytidine deaminases APOBEC3C and APOBEC3D promote DNA replication stress resistance in pancreatic cancer cells. Nat Cancer. 2024;5:895–915. https://doi.org/10.1038/s43018-024-00742-z
72. Stoof J, Harrold E, Mariottino S, Lowery MA, Walsh N. DNA damage repair deficiency in pancreatic ductal adenocarcinoma: preclinical models and clinical perspectives. Front Cell Dev Biol. 2021;9:749490. https://doi.org/10.3389/fcell.2021.749490
73. Ribeiro CF, Rodrigues S, Bastos DC, Fanelli GN, Pakula H, Foiani M, et al. Blocking lipid synthesis induces DNA damage in prostate cancer and increases cell death caused by PARP inhibition. Sci Signal. 2024;17:eadh1922. https://doi.org/10.1126/scisignal.adh1922
74. Qu S, Qi S, Zhang H, Li Z, Wang K, Zhu T, et al. Albumin-bound paclitaxel augments temozolomide treatment sensitivity of glioblastoma cells by disrupting DNA damage repair and promoting ferroptosis. J Exp Clin Cancer Res. 2023;42:285. https://doi.org/10.1186/s13046-023-02843-6
75. Chen H, Li Y, Li H, Chen X, Fu H, Mao D, et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature. 2024;631:663–669. https://doi.org/10.1038/s41586-024-07620-9
76. Chen Y, Wu J, Zhai L, Zhang T, Yin H, Gao H, et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell. 2024;187:294–311.e21. https://doi.org/10.1016/j.cell.2023.11.022
77. Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, et al. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. https://doi.org/10.1016/j.molcel.2016.06.020
78. Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology‑mediated end‑joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–237. https://doi.org/10.1038/nsmb.2961
79. Zatreanu D, Robinson HMR, Alkhatib O, Boursier M, Finch H, Geo L, et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat Commun. 2021;12:3636. https://doi.org/10.1038/s41467-021-23463-8
80. Blukacz L, Nuciforo S, Fucile G, Trulsson F, Duthaler U, Wieland S, et al. Inhibition of the transmembrane transporter ABCB1 overcomes resistance to doxorubicin in patient-derived organoid models of HCC. Hepatol Commun. 2024;8:e0000000000000437. https://doi.org/10.1097/HC9.0000000000000437
81. Nimmakayala RK, Leon F, Rachagani S, Rauth S, Nallasamy P, Marimuthu S, et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene. 2021;40:215–231. https://doi.org/10.1038/s41388-020-01518-2
82. Chau CH, Steeg PS, Figg WD. Antibody‑drug conjugates for cancer. Lancet. 2019;394:793–804. https://doi.org/10.1016/S0140-6736(19)31774-X
83. Benelli R, Costa D, Barboro P, Poggi A, Matis S, Zocchi MR, et al. Targeting colorectal cancer organoids with zoledronic acid conjugated to the anti-EGFR antibody cetuximab. J Immunother Cancer. 2022;10:e005660. https://doi.org/10.1136/jitc-2022-005660
84. Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389-1400. https://doi.org/10.1038/nm.3388
85. Cortés J, Kim SB, Chung WP, Im SA, Park YH, Hegg R, et al. Trastuzumab deruxtecan versus trastuzumab emtansine for breast cancer. N Engl J Med. 2022;386:1143–1154. https://doi.org/10.1056/NEJMoa2115022
86. Díaz‑Rodríguez E, Gandullo‑Sánchez L, Ocaña A, Pandiella A. Novel ADCs and strategies to overcome resistance to anti-HER2 ADCs. Cancers (Basel). 2021;14:154. https://doi.org/10.3390/cancers14010154
87. Wolf DM, Yau C, Wulfkuhle J, Brown‑Swigart L, Gallagher RI, Lee PRE, et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: predictive biomarkers across 10 cancer therapies. Cancer Cell. 2022;40:609–623.e6. https://doi.org/10.1016/j.ccell.2022.05.005
88. Chen YF, Zhang QH, Zhang ZW, Zhou YJ, Liu CC, Shao ZM, et al. SNX10 deficiency impairs sensitivity to anti-HER2 antibody–drug conjugates via altering HER2 trafficking in HER2-positive breast cancer. Proc Natl Acad Sci U S A. 2025;122:e2417586122. https://doi.org/10.1073/pnas.2417586122
89. Rathje F, Klingler S, Aberger F. Organoids for modeling (colorectal) cancer in a dish. Cancers (Basel). 2022;14:5416. https://doi.org/10.3390/cancers14215416
90. Tardito S, Matis S, Zocchi MR, Benelli R, Poggi A. Epidermal growth factor receptor targeting in colorectal carcinoma: antibodies and patient‑derived organoids as a smart model to study therapy resistance. Int J Mol Sci. 2024;25:7131. https://doi.org/10.3390/ijms25137131
91. Liu T, Wang H, Chen Y, Wan Z, Du Z, Shen H, et al. SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation. J Exp Clin Cancer Res. 2023;42(1):234. https://doi.org/10.1186/s13046-023-02789-9
92. Elurbide J, Colyn L, Latasa MU, Uriarte I, Mariani S, Lopez‑Pascual A, et al. Identification of PRMT5 as a therapeutic target in cholangiocarcinoma. Gut. 2024;74(1):116-127. https://doi.org/10.1136/gutjnl-2024-332998
93. Zhang T, Febres‑Aldana C, Liu Z, Dix JM, Cheng R, Dematteo RG, et al. HER2 antibody-drug conjugates are active against desmoplastic small round cell tumor. Clin Cancer Res. 2024;30:4701-4713. https://doi.org/10.1158/1078-0432.CCR-24-1835
94. Weng W, Meng T, Zhao Q, Shen Y, Fu G, Shi J, et al. Antibody-Exatecan conjugates with a novel self-immolative moiety overcome resistance in colon and lung cancer. Cancer Discov. 2023;13:950-973. https://doi.org/10.1158/2159-8290.CD-22-1368
95. Hong X, Chen X, Wang H, Xu Q, Xiao K, Zhang Y, et al. A HER2-targeted antibody-drug conjugate, RC48-ADC, exerted promising antitumor efficacy and safety with intravesical instillation in preclinical models of bladder cancer. Adv Sci (Weinh). 2023;10:e2302377. https://doi.org/10.1002/advs.202302377
96. Guo C, Figueiredo I, Gurel B, Neeb A, Seed G, Crespo M, et al. B7-H3 as a therapeutic target in advanced prostate cancer. Eur Urol. 2023;83:e168-e169. https://doi.org/10.1016/j.eururo.2022.09.004
97. Miranda Furtado CL, Dos Santos Luciano MC, Silva Da Santos R, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019;14:1164-1176. https://doi.org/10.1080/15592294.2019.1640546
98. Parizadeh SM, Jafarzadeh Esfehani R, Ghandehari M, Seifi S, Parizadeh SMR, Moetamani-Ahmadi M, et al. Epigenetic drug therapy in the treatment of colorectal cancer. Curr Pharm Des. 2018;24:2701-2709. https://doi.org/10.2174/1381612824666180730151904
99. Morgan AG, Griffin MF, Longaker MT, Norton JA. Precision medicine: IL-1RA and pancreatic cancer organoids. Biology (Basel). 2025;14:60604. https://doi.org/10.3390/biology14060604
100. Liu C, Li J, Xu F, Chen L, Ni M, Wu J, et al. PARP1-DOT1L transcription axis drives acquired resistance to PARP inhibitor in ovarian cancer. Mol Cancer. 2024;23:111. https://doi.org/10.1186/s12943-024-02025-8
101. Wang F, Yu X, Qian J, Cao Y, Dong S, Zhan S, et al. A novel SIK2 inhibitor SIC-19 exhibits synthetic lethality with PARP inhibitors in ovarian cancer. Drug Resist Updat. 2024;74:101077. https://doi.org/10.1016/j.drup.2024.101077
102. Spender LC, Watt DM, Baxter MA, Shuttleworth MK, Walker K, Savage AR, et al. AKT/mTOR as a targetable hub to overcome multimodal resistance to EGFR inhibitors in oesophageal squamous cell carcinoma. Br J Cancer. 2025;133(5):709-722. https://doi.org/10.1038/s41416-025-03093-3
103. Zhou Z, Van der Jeught K, Li Y, Sharma S, Yu T, Moulana I, et al. A T cell-engaging tumor organoid platform for pancreatic cancer immunotherapy. Adv Sci (Weinh). 2023;10:e2300548. https://doi.org/10.1002/advs.202300548
104. Xia Y, Sun M, Huang H, Jin WL. Drug repurposing for cancer therapy. Signal Transduct Target Ther. 2024;9(1):92. https://doi.org/10.1038/s41392-024-01808-1
105. Sada N, Lee S, Katsu T, Otsuki T, Inoue T. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science. 2015;347:1362-1367. https://doi.org/10.1126/science.aaa1299
106. Frampton JE. Stiripentol: a review in Dravet syndrome. Drugs. 2019;79:1785-1796. https://doi.org/10.1007/s40265-019-01204-y
107. Xie CK, Liao CY, Lin HY, Wu YD, Lu FC, Huang XX, et al. Sulindac (K-80003) with nab-paclitaxel and gemcitabine overcomes drug-resistant pancreatic cancer. Mol Cancer. 2024;23:215. https://doi.org/10.1186/s12943-024-02128-2
108. Suvilesh KN, Manjunath Y, Nussbaum YI, Gadelkarim M, Raju M, Srivastava A, et al. Targeting AKR1B10 by drug repurposing with epalrestat overcomes chemoresistance in non–small cell lung cancer patient-derived tumor organoids. Clin Cancer Res. 2024;30(17):3855-3867. https://doi.org/10.1158/1078-0432.CCR-23-3980
109. Yehya A, Ghamlouche F, Karami R, Hachem S, Salhab Z, Liu YN, et al. Repurposing piroxicam enhances the antineoplastic effects of docetaxel and enzalutamide in prostate cancer cells using 2D and 3D in vitro culture models. Front Cell Dev Biol. 2025;13:1551010. https://doi.org/10.3389/fcell.2025.1551010
110. Sun L, Kang X, Ju H, Wang C, Yang G, Wang R, et al. A human mucosal melanoma organoid platform for modeling tumor heterogeneity and exploring immunotherapy combination options. Sci Adv. 2023;9(43):eadg6686. https://doi.org/10.1126/sciadv.adg6686
111. Al Shihabi A, Tebon PJ, Nguyen HTL, Chantharasamee J, Sartini S, Davarifar A, et al. The landscape of drug sensitivity and resistance in sarcoma. Cell Stem Cell. 2024;31:1524-1542.e1524. https://doi.org/10.1016/j.stem.2024.08.010
112. Kim SY, de Weert TAE, Vermeulen M, Ringnalda F, Kester L, Zsiros J, et al. Organoid drug profiling identifies methotrexate as a therapy for SCCOHT, a rare pediatric cancer. Sci Adv. 2025;11:eadq1724. https://doi.org/10.1126/sciadv.adq1724
113. Verstegen MMA, Coppes RP, Beghin A, De Coppi P, Gerli MFM, de Graeff N, et al. Clinical applications of human organoids. Nat Med. 2025;31:409-421. https://doi.org/10.1038/s41591-024-03489-3
114. Zhao Y, Li S, Zhu L, Huang M, Xie Y, Song X, et al. Personalized drug screening using patient-derived organoid and its clinical relevance in gastric cancer. Cell Rep Med. 2024;5:101627. https://doi.org/10.1016/j.xcrm.2024.101627
115. Jensen LH, Rogatto SR, Lindebjerg J, Havelund B, Abildgaard C, do Canto LM, et al. Precision medicine applied to metastatic colorectal cancer using tumor-derived organoids and in-vitro sensitivity testing: a phase 2, single-center, open-label, and non-comparative study. J Exp Clin Cancer Res. 2023;42:115. https://doi.org/10.1186/s13046-023-02683-4
116. Ma YS, Yang XL, Xin R, Wu TM, Shi Y, Zhang DD, et al. The power and the promise of organoid models for cancer precision medicine with next-generation functional diagnostics and pharmaceutical exploitation. Transl Oncol.2021;14:101126. https://doi.org/10.1016/j.tranon.2021.101126
117. van de Haar J, Ma X, Ooft SN, van der Helm PW, Hoes LR, Mainardi S, et al. Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat Med. 2023;29:605-614. https://doi.org/10.1038/s41591-023-02240-8
118. Hedayat S, Cascione L, Cunningham D, Schirripa M, Lampis A, Hahne JC, et al. Circulating microRNA analysis in a prospective co-clinical trial identifies MIR652-3p as a response biomarker and driver of regorafenib resistance mechanisms in colorectal cancer. Clin Cancer Res. 2024;30:2140-2159. https://doi.org/10.1158/1078-0432.ccr-23-2748
119. Mayer RJ, Van Cutsem E, Falcone A, Yoshino T, Garcia-Carbonero R, Mizunuma N, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372:1909-1919. https://doi.org/10.1056/NEJMoa1414325
120. Van Cutsem E, Mayer RJ, Laurent S, Winkler R, Grávalos C, Benavides M, et al. The subgroups of the phase III RECOURSE trial of trifluridine/tipiracil (TAS-102) versus placebo with best supportive care in patients with metastatic colorectal cancer. Eur J Cancer. 2018;90:63-72. https://doi.org/10.1016/j.ejca.2017.10.009
121. Beltran H, Oromendia C, Danila DC, Montgomery B, Hoimes C, Szmulewitz RZ, et al. A phase II trial of the Aurora kinase A inhibitor alisertib for patients with castration-resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin Cancer Res. 2019;25:43-51. https://doi.org/10.1158/1078-0432.ccr-18-1912
122. Goodwin CM, Waters AM, Klomp JE, Javaid S, Bryant KL, Stalnecker C, et al. Combination therapies with CDK4/6 inhibitors to treat KRAS-mutant pancreatic cancer. Cancer Res. 2023;83:141-157. https://doi.org/10.1158/0008-5472.can-22-0391
123. Xu H, Jiao D, Liu A, Wu K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol. 2022;15:58. https://doi.org/10.1186/s13045-022-01278-4
124. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373-386.e310. https://doi.org/10.1016/j.cell.2017.11.010
125. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol.2020;21:571-584. https://doi.org/10.1038/s41580-020-0259-3
126. Koh V, Chakrabarti J, Torvund M, Steele N, Hawkins JA, Ito Y, et al. Hedgehog transcriptional effector GLI mediates mTOR-induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021;518:59-71. https://doi.org/10.1016/j.canlet.2021.06.007
127. Holokai L, Chakrabarti J, Lundy J, Croagh D, Adhikary P, Richards SS, et al. Murine- and human-derived autologous organoid/immune cell co-cultures as pre-clinical models of pancreatic ductal adenocarcinoma. Cancers (Basel). 2020;12:3816. https://doi.org/10.3390/cancers12123816
128. Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10:1129-1139. https://doi.org/10.1158/2159-8290.cd-20-0187
129. Wang K, Yuen ST, Xu J, Lee SP, Yan HHN, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. https://doi.org/10.1038/ng.2983
130. Zhao Y, Li ZX, Zhu YJ, Fu J, Zhao XF, Zhang YN, et al. Single-cell transcriptome analysis uncovers intratumoral heterogeneity and underlying mechanisms for drug resistance in hepatobiliary tumor organoids. Adv Sci (Weinh). 2021;8:e2003897. https://doi.org/10.1002/advs.202003897
131. Wang G, Xu Y, Wang Q, Chai Y, Sun X, Yang F, et al. Rare and undiagnosed diseases: from disease-causing gene identification to mechanism elucidation. Fundam Res. 2022;2:918-928. https://doi.org/10.1016/j.fmre.2022.09.002
132. Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515-528.e517. https://doi.org/10.1016/j.cell.2018.03.017
133. Maru Y, Tanaka N, Itami M, Hippo Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol Oncol. 2019;154:189-198. https://doi.org/10.1016/j.ygyno.2019.05.005
134. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández‑Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920-926. https://doi.org/10.1126/science.aao2774
135. Gronholm M, Feodoroff M, Antignani G, Martins B, Hamdan F, Cerullo V. Patient-derived organoids for precision cancer immunotherapy. Cancer Res. 2021;81:3149-3155. https://doi.org/10.1158/0008-5472.can-20-4026
136. Zhou X, Qu M, Tebon P, Jiang X, Wang C, Xue Y, et al. Screening cancer immunotherapy: when engineering approaches meet artificial intelligence. Adv Sci (Weinh). 2020;7:2001447. https://doi.org/10.1002/advs.202001447
137. Huang Y, Huang Z, Tang Z, Chen Y, Huang M, Liu H, et al. Research progress, challenges, and breakthroughs of organoids as disease models. Front Cell Dev Biol. 2021;9:740574. https://doi.org/10.3389/fcell.2021.740574
138. Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653-658. https://doi.org/10.1016/j.stem.2013.11.002
139. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407-418. https://doi.org/10.1038/s41568-018-0007-6
140. De Sousa E Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676-680. https://doi.org/10.1038/nature21713
141. Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8:745-760. https://doi.org/10.15252/emmm.201606184
142. Michels BE, Mosa MH, Streibl BI, Zhan T, Menche C, Abou‑El‑Ardat K, et al. Pooled in vitro and in vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell. 2020;26:782-792.e787. https://doi.org/10.1016/j.stem.2020.04.003
143. Yang J, Huang S, Cheng S, Jin Y, Zhang N, Wang Y. Application of ovarian cancer organoids in precision medicine: key challenges and current opportunities. Front Cell Dev Biol. 2021;9:701429. https://doi.org/10.3389/fcell.2021.701429
144. Liang J, Wei J, Cao J, Qian J, Gao R, Li X, et al. In-organoid single-cell CRISPR screening reveals determinants of hepatocyte differentiation and maturation. Genome Biol. 2023;24:251. https://doi.org/10.1186/s13059-023-03084-8
145. Guo D, Yao B, Shao WW, Zuo JC, Chang ZH, Shi JX, et al. The critical role of YAP/BMP/ID1 axis on simulated microgravity-induced neural tube defects in human brain organoids. Adv Sci (Weinh). 2025;12:e2410188. https://doi.org/10.1002/advs.202410188
146. Bai L, Wu Y, Li G, Zhang W, Zhang H, Su J. AI-enabled organoids: construction, analysis, and application. Bioact Mater. 2024;31:525-548. https://doi.org/10.1016/j.bioactmat.2023.09.005
147. Man Y, Liu Y, Chen Q, Zhang Z, Li M, Xu L, et al. Organoids-on-a-chip for personalized precision medicine. Adv Healthc Mater. 2024;13:e2401843. https://doi.org/10.1002/adhm.202401843
148. Wahida A, Buschhorn L, Frohling S, Jost PJ, Schneeweiss A, Lichter P, et al. The coming decade in precision oncology: six riddles. Nat Rev Cancer. 2023;23:43-54. https://doi.org/10.1038/s41568-022-00529-3
149. Johansson A, Andreassen OA, Brunak S, Franks PW, Hedman H, Loos RJF, et al. Precision medicine in complex diseases - molecular subgrouping for improved prediction and treatment stratification. J Intern Med. 2023;294:378-396. https://doi.org/10.1111/joim.13640
150. Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40:920-938. https://doi.org/10.1016/j.ccell.2022.08.011
151. Deng S, Li C, Cao J, Cui Z, Du J, Fu Z, et al. Organ-on-a-chip meets artificial intelligence in drug evaluation. Theranostics. 2023;13:4526-4558. https://doi.org/10.7150/thno.87266
152. Liu J, Wang Q, Le Y, Hu M, Li C, An N, et al. 3D-bioprinting for precision microtissue engineering: advances, applications, and prospects. Adv Healthc Mater. 2025;14:e2403781. https://doi.org/10.1002/adhm.202403781
153. Shukla P, Yeleswarapu S, Heinrich MA, Prakash J, Pati F. Mimicking tumor microenvironment by 3D bioprinting: 3D cancer modeling. Biofabrication. 2022;14. https://doi.org/10.1088/1758-5090/ac6d11
154. Lawlor KT, Vanslambrouck JM, Higgins JW, Chambon A, Bishard K, Arndt D, et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat Mater. 2021;20:260-271. https://doi.org/10.1038/s41563-020-00853-9
155. Shukla P, Bera AK, Ghosh A, Kiranmai G, Pati F. Assessment and process optimization of high throughput biofabrication of immunocompetent breast cancer model for drug screening applications. Biofabrication. 2024;16. https://doi.org/10.1088/1758-5090/ad586b
156. Subtil B, Iyer KK, Poel D, Bakkerus L, Gorris MAJ, Cuenca Escalona J, et al. Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front Immunol. 2023;14:1105244. https://doi.org/10.3389/fimmu.2023.1105244
157. Magré L, Verstegen MMA, Buschow S, van der Laan LJW, Peppelenbosch M, Desai J. Emerging organoid–immune co-culture models for cancer research: from oncoimmunology to personalized immunotherapies. J Immunother Cancer. 2023;11:6290. https://doi.org/10.1136/jitc-2022-006290
158. Yang R, Yu Y. Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 2023;562:216180. https://doi.org/10.1016/j.canlet.2023.216180
159. Saorin G, Caligiuri I, Rizzolio F. Microfluidic organoids-on-a-chip: the future of human models. Semin Cell Dev Biol.2023;144:41-54. https://doi.org/10.1016/j.semcdb.2022.10.001
160. Lee JW, Hur J, Kwon YW, Chae CW, Choi JI, Hwang I, et al. KAI1 (CD82) is a key molecule to control angiogenesis and switch angiogenic milieu to quiescent state. J Hematol Oncol. 2021;14:148. https://doi.org/10.1186/s13045-021-01147-6
161. Luo Q, Wang J, Zhao W, Peng Z, Liu X, Li B, et al. Vasculogenic mimicry in carcinogenesis and clinical applications. J Hematol Oncol. 2020;13:19. https://doi.org/10.1186/s13045-020-00858-6
162. Quintard C, Tubbs E, Jonsson G, Jiao J, Wang J, Werschler N, et al. A microfluidic platform integrating functional vascularized organoids-on-chip. Nat Commun. 2024;15:1452. https://doi.org/10.1038/s41467-024-45710-4
163. Du Y, Wang YR, Bao QY, Xu XX, Xu C, Wang S, et al. Personalized vascularized tumor organoid-on-a-chip for tumor metastasis and therapeutic targeting assessment. Adv Mater. 2025;37:e2412815. https://doi.org/10.1002/adma.202412815
Declarations
Funding Statement: This work was funded by Chongqing Science and Technology Bureau (Grant No. cstc2022jxjl20039), Talent Program of Chongqing (Grants No. cstc2024rcih-bgzxm0162, YXGD202403), Chongqing Health Commission (Grant No. 2023ZDXM029 and 2023MSXM043), Scientific and Technological Research Program of Chongqing Municipal Education Commission (Grant No. KJQN20230013), Project for Enhancing Scientific Research Capabilities of Chongqing University Cancer Hospital (Grant No. 2023nlts009), Beijing Health Alliance Charitable Foundation (Grant No. BJHA-CRP-089), and Chongqing Shapingba District Health Commission (Grant No. 2023SQKWLH021).
Declaration of Competing Interest: The authors declare no financial or personal relationships with other individuals or organizations that could inappropriately influence or bias the content of this work. All authors have read the final version of the manuscript and confirm that there are no competing interests.
Consent for publication: All authors have approved the final version of the manuscript.
Use of Artificial Intelligence (AI) Disclosure: This article was written by human contributors. Artificial intelligence-based tools were used to improve grammar, language, and readability, without affecting the scientific content, data interpretation, or conclusions of the article. The authors reviewed and verified all content to ensure its accuracy and integrity.
Dual Use Research of Concern (DURC): “The authors confirm that this research does not constitute DURC as defined by the U.S. Government Policy for Oversight of Life Sciences DURC.”
Data Availability Statements (DAS): “No datasets were generated or analyzed in the current study.”
Ethics approval and consent to participate: Not applicable, as this study did not involve the conduct of research.
Authors’ affiliations
1. Department of Gynecologic Oncology, Chongqing University Cancer Hospital & Chongqing Cancer Institute & Chongqing Cancer Hospital, Chongqing, China.
2. Chongqing Specialized Medical Research Center of Ovarian Cancer, Chongqing, China.
3. Organoid Transformational Research Center, Chongqing Key Laboratory for the Mechanism and Intervention of Cancer Metastasis, Chongqing University Cancer Hospital, Chongqing, China.
* LW, QT, and LZ contributed equally to this work and are designated as co-first authors.
# Correspondence: Dongling Zou: Email: zoudl_cqch@cqu.edu.cn and Haixia Wang: Email: wanghx1985@126.com
CRediT authorship contribution statement: LW reviewed the collected papers and created the figures/tables. LW, QT, and LZ originally drafted the manuscript. MSH, XPZ, QZ, and LL collected literature on resistance. HXW and DLZ initiated the study and revised the manuscript. All authors contributed to the work and approved the final version of the manuscript.
ORCID ID:
Dongling Zou: https://orcid.org/0000-0001-9158-8783
Haixia Wang: https://orcid.org/0000-0002-4847-1421
How to cite
Wang L, Tang Q, Zhong L, He M, Zhu X, Zheng Q, et al. Organoids as Next-Generation Models of Tumor Drug Resistance. Cancer Biome and Targeted Therapy. Published online 2025 Dec.