REVIEW ARTICLE OPEN ACCESS
The Cancer Microbiome: Mechanistic and Translational Insights into Oncogenesis and Therapy
Yangyun Wang1*, Youyang Shi2*, Jingjing Duan3*, Haijia Tang2, Lei Chen3#, Sheng Liu2#, Jianfeng Yang3#
Received 2025 Oct 2
Accepted 2025 Nov 3
Epub ahead of print: December 2025
Published in issue 2026 Feb 15
Correspondence: Jianfeng Yang - Email: 0156333@163.com
Sheng Liu - Email: lslhtcm@163.com
Chen Lei - Email: joe8989@163.com
The author’s information is available at the end of the article.
© 2026 The Author(s). Published by GCINC Press. Open Access licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. To view a copy: https://creativecommons.org/licenses/by/4.0/
Abstract
The human microbiome, including bacteria, fungi, and viruses, is increasingly recognized as a key player in cancer development and progression. Established oncogenic microorganisms such as Helicobacter pylori, human papillomavirus, and hepatitis viruses account for nearly 15% of cancers worldwide. Recent sequencing studies have further revealed the presence of diverse microbial communities in organs previously thought to be sterile. Microbial dysbiosis can promote carcinogenesis through multiple mechanisms, including DNA damage and genomic instability, chronic inflammation, immune suppression, and metabolic reprogramming. Distinct microbial signatures have been identified across various central malignancies, including lung, oral, gastric, pancreatic, colorectal, hepatocellular, breast, prostate, and gynecological cancers, highlighting their potential for both diagnostic and prognostic applications. Moreover, modulation of the microbiome is emerging as a promising therapeutic strategy, with applications ranging from probiotics and prebiotics to enhancing responses to immunotherapy, chemotherapy, and fecal microbiota transplantation. This review synthesizes current knowledge of microbiome-cancer interactions, emphasizes their translational implications, and outlines future directions for leveraging the microbiome in precision oncology.
Keywords: Cancer microbiome, Tumor microenvironment, Microbial dysbiosis, Drug resistance, Microbiota, Biofilms, Host-microbe interactions.
1. Introduction
Cancer remains the second leading cause of mortality worldwide. While conventional paradigms have long attributed carcinogenesis primarily to genetic predisposition and environmental exposures, mounting evidence now illuminates a pivotal role for the microbiome in tumor initiation and progression.
Table 1: Cancer types and key microbial associations.
| Cancer Type | Associated Microbes | Mechanisms | Clinical Relevance |
|---|---|---|---|
| Lung | Veillonella (3-5), Fusobacterium (6), Akkermansia (7, 8) | Inflammation, immune modulation | Potential biomarker |
| Oral | Porphyromonas gingivalis (9), Fusobacterium nucleatum (10) | EMT, immune suppression | Diagnostic saliva tests |
| Gastric | Helicobacter pylori (11, 12), Candida albicans (13), EBV (14) | DNA damage, chronic gastritis, PD-L1 upregulation | Target for eradication therapy |
| Pancreatic | Malassezia, P. (15) gingivalis (16), Fusobacterium (17) | Complement activation, immune suppression | Prognostic biomarkers |
| Colorectal |
Fusobacterium nucleatum (18), Bacteroides fragilis (19), pks+ E. coli (20) |
DNA alkylation, T cell inhibition | Stool-based screening |
| Hepatocellular Carcinoma | Dysbiotic gut flora (21) | Bile acid metabolism, gut–liver axis | Microbiome–liver cancer therapy |
| Breast | Lactobacillus (22) | Estrogen metabolism, immune dysfunction | Tumor microbiome studies |
In this context, specific microbial infections, including those caused by viruses, bacteria, and fungi, are increasingly recognized as significant risk factors for cancer development. Epidemiological data indicate that approximately 15% of cancers globally can be attributed to infection with carcinogenic microbes, with this burden disproportionately affecting low and middle-income countries (1). Moreover, co-infection with multiple microbial agents may synergistically amplify the likelihood of cancer development. Notable contributors to this global cancer burden include Helicobacter pylori, human papillomavirus (HPV), hepatitis B and C viruses (HBV and HCV), Epstein-Barr virus (EBV), human immunodeficiency virus (HIV), and human herpesviruses (HHV), each contributing to varying extents. These infection-associated cancers underscore the oncogenic potential of specific microbes. Importantly, accumulating evidence suggests that cancer risk is not limited to direct infection alone. Advances in sequencing and microbial ecology have revealed that the broader commensal microbiome, encompassing both classical pathogens and other microbes, also influences the tumor microenvironment, modulates immune surveillance, and impacts cancer-related processes. This paradigm shift has thus expanded the focus from isolated infectious microbes to the complex microbial communities that coexist within the human host.
The human microbiome includes all microbial communities residing on and within the human body. It is intricately linked to multiple facets of host health and disease (2). Microbial ecosystems exist across virtually all examined human ecological niches. This includes the oral cavity, cutaneous surfaces, gastrointestinal tract, esophagus, lungs, and beyond (Figure 1).
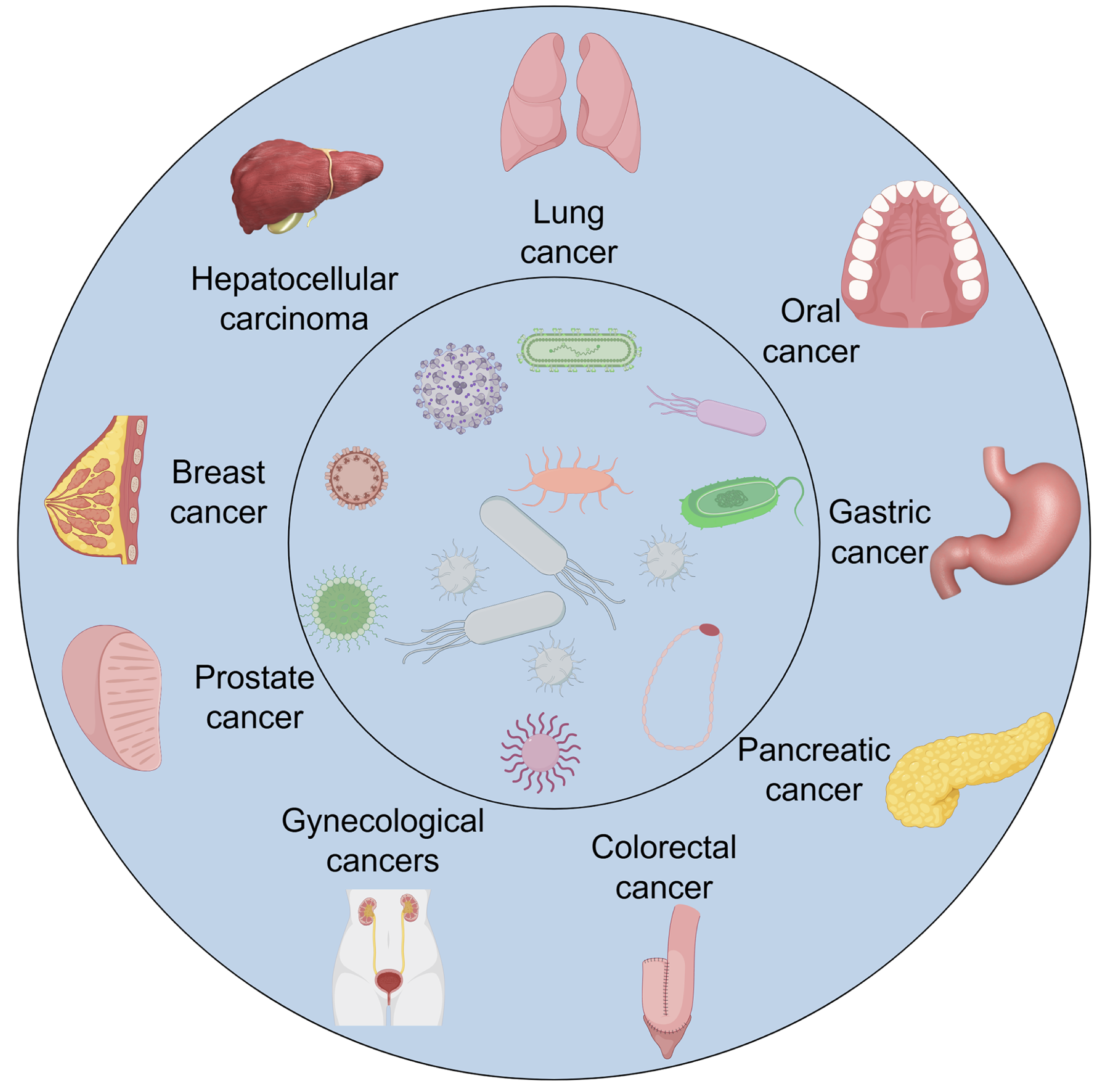
Figure 1. Overview of the human microbiome and its interactions with the host. The figure provides a schematic overview of the human microbiome across major anatomical sites, including the gastrointestinal tract, oral cavity, respiratory system, urogenital tract, and skin. Each region harbors a distinct microbial community composed of bacteria, fungi, and viruses that work collectively to maintain host homeostasis. Microbiome–host interactions occur through immune signaling, metabolic crosstalk, and regulation of epithelial barrier integrity, supporting both local and systemic physiological balance. The diagram also highlights how disruptions in these interactions, referred to as dysbiosis, can lead to immune imbalance, chronic inflammation, and increased susceptibility to disease, including various cancers.
These complex microbiotas comprise diverse microorganisms, including bacteria, archaea, viruses, bacteriophages, and fungi. Together, they shape the ever-changing microbial environment of the human body. Disruptions in the gut microbial balance, often referred to as dysbiosis, are increasingly associated with tumor development. Gastric cancer is a clear example of the connection between microbial imbalance and host epithelial behavior. In addition to the well-known role of Helicobacter pylori, acid-tolerant bacteria, such as Lactobacillus, Veillonella, and Clostridium species, have been observed to increase in the stomach. This shift suggests their potential role in cancer development when the microbial balance is disrupted. In lung cancer, distinct microbial signatures associate with specific histological subtypes. Small-cell lung cancer (SCLC) is associated with increased prevalence of Kl, Acidovorax, Polaromonas, Rhodoferax, and Xylobacter. In contrast, non-small cell lung cancer (NSCLC) patients have elevated Ruminococcus spp., Akkermansia muciniphila, Eubacterium spp., and Alistipes spp. These differential patterns suggest that respiratory and gut microbiota may both contribute to lung cancer pathophysiology. This highlights the potential utility of the microbiome as a biomarker or therapeutic target in oncological management.
This review consolidates recent insights into the role of the microbiome in major cancer types, including lung, oral, pancreatic, gastric, colorectal, hepatocellular, breast, prostate, and gynecological cancers. The review connects the mechanistic bases discussed above with prospective clinical strategies. It also highlights future directions for translational research in this rapidly evolving field.
2. The link between microbiome and cancer development
The microbiome contributes to oncogenesis by inducing genomic instability and structural aberrations (23). Within the tumor microenvironment (TME), certain microorganisms and their secreted toxins can directly damage host DNA, thereby increasing the mutational burden in colonized tissues. As DNA lesions accumulate beyond a critical threshold, regulatory networks governing cellular proliferation become disrupted, ultimately driving tumor initiation and progression (24, 25). A well-characterized example is Escherichia coli strains harboring the polyketide synthase (pks) gene cluster (pks⁺E. coli), which has been implicated in colorectal carcinogenesis by inducing somatic mutations and DNA breaks (26, 27) (Figure 2A).
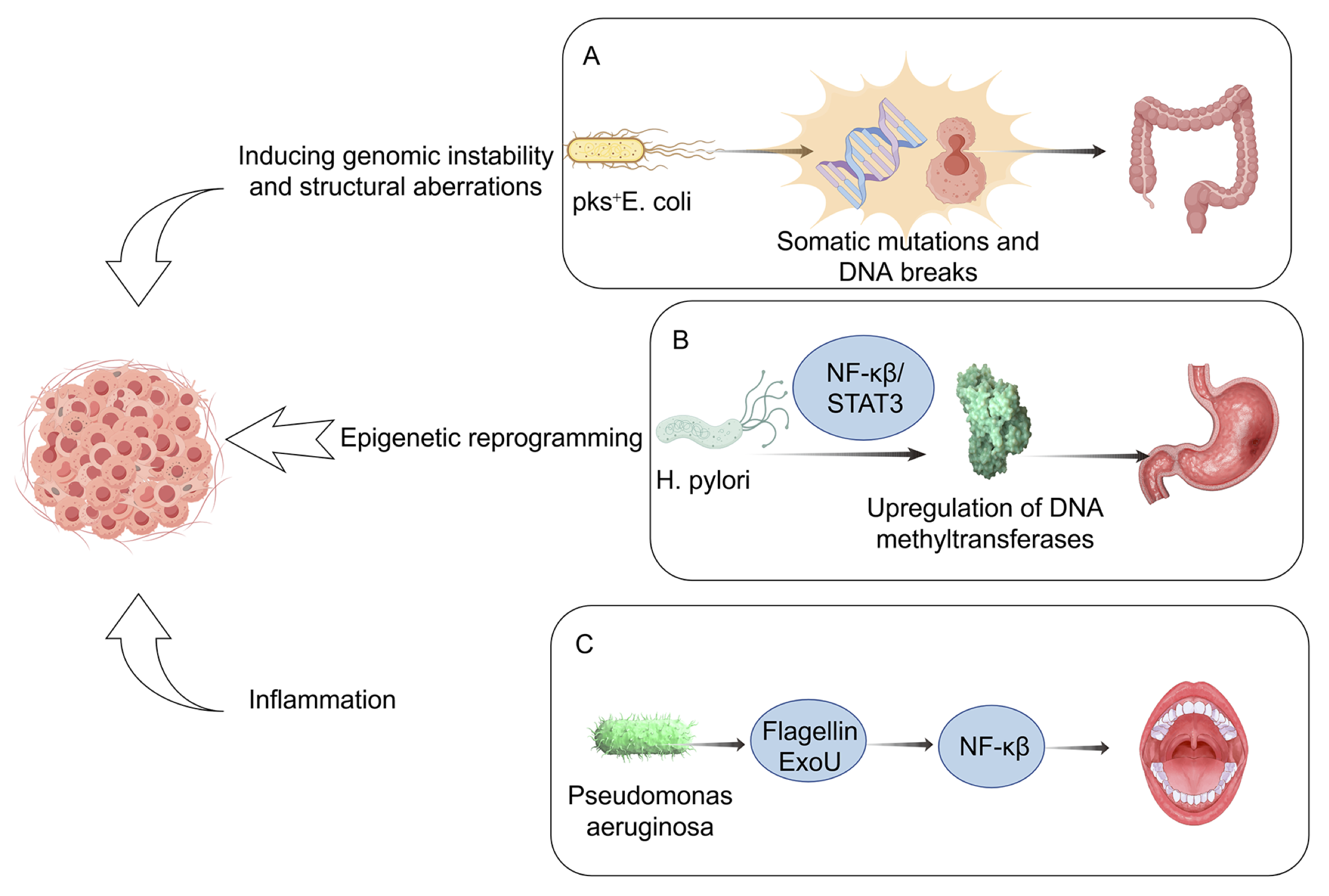
Figure 2. Mechanistic pathways linking the microbiome to cancer development. This schematic summarizes the principal mechanisms through which microbial dysbiosis promotes carcinogenesis. Specific taxa or microbial products may induce DNA damage and genomic instability, trigger chronic inflammation, alter host immune surveillance, and reprogram tumor metabolism. Arrows indicate directional interactions among microbial, immune, and epithelial compartments. The illustration highlights how both commensal and pathogenic microbes can shape the tumor microenvironment, supporting cancer initiation and progression.
In a seminal study, Pleguezuelos-Manzano et al. co-cultured human intestinal organoids derived from healthy stem cells with pks⁺ E. coli, demonstrating that long-term bacterial exposure induces distinctive mutational signatures, including single base substitutions (SBS-pks) and small insertion-deletion events (ID-pks). Beyond bacterial genotoxins, microbial metabolites play a decisive role in promoting DNA damage. For instance, small-molecule derivatives from diverse gut microbiota directly impair DNA integrity in acellular assays, induce double-strand break (DSB) markers (γ-H2AX), and cause epithelial cell-cycle arrest. Specifically, indolimine metabolites from Morganella morganii have been shown to exacerbate colon tumorigenesis in germ-free mice (28). Additional pathogenic mechanisms involve effector proteins secreted by enteropathogenic E. coli and Helicobacter pylori that disrupt DNA mismatch repair, thereby destabilizing the genome and fueling tumorigenesis (29, 30). These processes are frequently associated with the overproduction of reactive oxygen species (ROS), hydrogen sulfide, and nitric oxide, molecules well known to contribute to genotoxic stress (31, 32). Viruses also represent important microbial drivers of genomic instability. For example, Merkel cell polyomavirus (MCPyV) integrates viral DNA into host genomes and persistently expresses the viral T antigen, which contributes to tumorigenesis in approximately 60% of Merkel cell carcinoma cases (33, 34). Beyond genetic alterations, microorganisms exert oncogenic influence through epigenetic reprogramming. H. pylori, particularly CagA-positive strains, can induce aberrant DNA hypermethylation of tumor suppressor gene promoters in gastric mucosa via nuclear factor kappa B (NF-κB)/STAT3-mediated upregulation of DNA methyltransferases. Microbe-driven epigenetic modifications, encompassing altered DNA methylation, dysregulated noncoding RNAs, and histone modifications, represent key mechanisms linking chronic infection to malignant transformation (Figure 2B).
Chronic inflammation and the sustained production of inflammatory mediators generate a tumor-permissive microenvironment, thereby constituting a major driver of carcinogenesis (35). Microbial components can act as key inflammatory triggers; for instance, Pseudomonas aeruginosa-derived factors such as flagellin and the cytotoxin ExoU exhibit strong pro-inflammatory activity by recruiting neutrophils and activating NF-κB signaling, ultimately accelerating the progression of oral cancer (36) (Figure 2C). Similarly, disruption of the intestinal epithelial barrier enables microbial products to translocate into host tissues, where they activate tumor-infiltrating dendritic cells (DCs) with inflammatory phenotypes. This, in turn, promotes the polarization of γδ T17 cells, which secrete high levels of pro-inflammatory cytokines, including IL-17, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and TNF-α. These mediators not only perpetuate inflammation but also recruit polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), thereby reprogramming the inflammatory milieu into an immunosuppressive state that facilitates colorectal cancer progression (37). As illustrated in Figure 2, the microbiome influences tumorigenesis through multiple converging axes, including genomic instability, chronic inflammation, immune suppression, and metabolic reprogramming. These pathways interact dynamically within the tumor microenvironment, underscoring the multifactorial nature of microbiome-driven carcinogenesis. The following sections present major cancer types in an order reflecting the progressive spectrum of microbial exposure and research development, from external or mucosal interfaces (e.g., lung and oral cavity) to internal organ systems (e.g., gastrointestinal, hepatic, and endocrine-related malignancies). The sequence aims to illustrate the expanding conceptual framework of microbiome-associated carcinogenesis.
2.1. Lung cancer
Lung cancer ranks as the foremost cause of cancer-related mortality worldwide and represents the second most frequently diagnosed malignancy, representing one of the most frequently diagnosed malignancies (38). The disease is often detected at advanced stages, and its etiology is predominantly linked to tobacco smoking. Nevertheless, epidemiological evidence indicates a rising incidence among never-smokers, now accounting for approximately 25% of cases (39). Biologically, lung cancer is a heterogeneous entity encompassing multiple histopathological subtypes, which are broadly categorized into two principal groups: small-cell lung carcinoma (SCLC), the most aggressive and lethal form, and non-small-cell lung carcinoma (NSCLC) (40). While SCLC accounts for 10-15% of lung cancer cases, approximately 85% are classified as NSCLC, which encompasses three predominant histological subtypes: adenocarcinoma, squamous cell carcinoma (SCC), and large-cell carcinoma, each characterized by distinct histological and molecular profiles. At the molecular level, the genomic architecture and genetic heterogeneity of lung cancer have been extensively delineated, underscoring its nature as a highly heterogeneous group of malignancies. Although recent advances have led to the development of targeted therapies for certain genetic subtypes, the overall survival rate for lung cancer remains alarmingly low. Cigarette smoking remains the primary risk factor, while other well-established contributors include ambient air pollution and occupational exposure to radon and asbestos (41, 42). As the mucosal organ with the largest surface area (e.g., upper vs. lower lobe) and a principal interface between the host and the external environment, the lung is uniquely positioned for continual exposure to airborne microorganisms and environmental pollutants (43,44). However, the precise mechanisms by which these environmental risk factors and other tumor-extrinsic influences drive lung carcinogenesis remain incompletely understood. Traditionally, healthy lungs were thought to be sterile; however, with the advent of increasingly sophisticated detection methodologies, including computed tomography (CT) imaging, polymerase chain reaction (PCR) assays, and 16S rRNA gene sequencing, investigations into the pulmonary microbiome have expanded considerably (45, 46). Since 2011, growing evidence has highlighted associations between distinct microbial communities and a range of pulmonary pathologies, confirming that they contain a variety of microorganisms (43, 47) (Table 2).
Table 2. Lung microbiome and its association with lung cancer.
| Lung microbiome | Types | Potential mechanisms |
|---|---|---|
| Pseudomonas (47, 49, 57-59) |
Gram-negative, aerobes |
These microbial alterations were found to correlate positively with macrophage abundance and elevated IFN-γ levels in bronchoalveolar lavage fluid, as well as with increased neutrophil elastase activity (66, 67). |
| Streptococcus (47, 60-64) |
Gram-positive, facultative anaerobes |
These microbes were shown to upregulate the ERK and PI3K signaling pathways, while exhibiting a negative correlation with active neutrophil elastase levels (68, 69). |
| Sphingomonas (49, 57, 65) |
Gram-negative, strictly aerobes |
They exhibited a positive correlation with macrophage abundance and with IFN-γ levels in the bronchoalveolar lavage (BAL)(66). |
| Propionibacterium |
Gram-positive, facultative anaerobes |
Not Described. |
| Acidovorax |
Gram-negative, aerobes |
Not Described. |
Compositional analyses of the pulmonary microbiome indicate a taxonomic architecture predominantly shaped by the phyla Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. Within this, the airway microbial repertoire encompasses diverse genera, including Prevotella, Veillonella, Streptococcus, Neisseria, Haemophilus, Fusobacterium, Sphingomonas, Pseudomonas, Acinetobacter, Megasphaera, Staphylococcus, and Corynebacterium, which contribute to the ecological complexity and potential functional interactions within the respiratory niche (47-49). Interestingly, marked compositional distinctions exist between the microbiota of the upper and lower respiratory tracts. In healthy individuals, the lower airways are predominantly colonized by Veillonella, Prevotella, and Streptococcus, accompanied by additional taxa such as Fusobacterium and Haemophilus, populations largely derived from the oral microbiome. Mounting evidence underscores the regulatory influence of the gut-lung axis, that is, the gut microbiota modulates pulmonary physiology and immune homeostasis. The intestinal microbiota, comprising a vast array of microbial species, exerts systemic effects on pulmonary immunity through the release of metabolites, microbial ligands, and immune mediators that circulate via the bloodstream. These products not only influence immune activity in the lungs but may also contribute to shaping the composition of the pulmonary microbiome. Conversely, the pulmonary microbial community plays a pivotal role in maintaining respiratory immune homeostasis, engaging in dynamic crosstalk with epithelial and immune cells to orchestrate both innate and adaptive immune responses (50, 51). Recent research has implicated the gut microbiome as a potential mediator linking these environmental exposures to lung tumorigenesis, suggesting that microbial dysbiosis may act in concert with chemical carcinogens to influence disease initiation and progression. Accumulating evidence underscores a strong association between dysbiosis of the gut microbiota and lung cancer. Liu et al. reported reduced microbial diversity and ecosystem stability in lung cancer patients, characterized by the enrichment of opportunistic pathogens and depletion of beneficial taxa (52). Zhuang et al. reported elevated Enterococcus abundance in the gut of lung cancer patients, alongside an overall decline in microbial functionality, suggesting that Enterococcus and Bifidobacterium may serve as potential biomarkers (53). Consistent with this, Zhang et al. observed reduced levels of Kluyvera, Escherichia-Shigella, Dialister, Faecalibacterium, and Enterobacter in lung cancer patients, while Veillonella, Fusobacterium, and Bacteroides were significantly enriched (54). Dysregulation of butyrate-producing bacteria has also been implicated: Gui et al. identified marked reductions in Clostridium leptum, Faecalibacterium prausnitzii, Ruminococcus, and Clostridial cluster I spp., whereas Eubacterium rectale and Clostridial cluster XIVa remained unaffected (55). Notably, elevated levels of Bacillus and Akkermansia muciniphila were associated with lung cancer progression (56).
In the context of lung malignancies, distinct microbial signatures have been documented. Accumulating evidence indicates that the pulmonary microbiota can remodel the local immune microenvironment, thereby contributing to tumor progression. In an autochthonous mouse model, Jin et al. provided compelling evidence that crosstalk between the lung microbiota and the host immune system is a critical driver of inflammatory signaling and lung tumorigenesis. They reported that tumor-bearing lungs harbored a distinct microbial signature, characterized by enrichment of taxa such as Herbaspirillum and Sphingomonadaceae. In contrast, healthy lungs were characterized by enrichment of Aggregatibacter and Lactobacillus. Elevated bacterial load and compositional shifts activated Myd88-dependent signaling in myeloid cells, triggering the secretion of IL-1β and IL-23. These cytokines, in turn, expanded and activated Vy6+Vδ1+γδ T cells, which produced IL-17 to amplify inflammation, while concurrently secreting IL-22 and other effector molecules that enhanced tumor cell proliferation. Notably, both germ-free and antibiotic-treated mice exhibited attenuated tumor progression, underscoring that commensal bacteria play an active role in facilitating lung carcinogenesis (70). Small cell lung cancer (SCLC) has been frequently associated with genera such as Klebsiella, Acidovorax, Polaromonas, Rhodoferax, Xylobacter, Eufluobacter, and Clostridium. In contrast, Prevotella and Pseudobutyrivibrio ruminis appear inversely correlated with the disease.
Conversely, non-small cell lung cancer (NSCLC) has been associated with increased relative abundance of Ruminococcus spp., Akkermansia muciniphila, Eubacterium spp., and Alistipes spp., and reduced prevalence of Bifidobacterium longum, Bifidobacterium adolescentis, and Parabacteroides distasonis. Collectively, these findings suggest that the gut microbiome may exert clinically relevant influences on lung cancer pathogenesis and progression. Although several taxa have been linked to lung cancer, findings are not always consistent across populations, which may reflect differences in diet, geography, and sequencing methods.
2.2. Oral cancer
The origins of oral microbiology can be traced to 1670, when Antonie van Leeuwenhoek, employing a microscope of his own design, first documented the presence of bacteria within the human oral cavity. His meticulous sketches and descriptions of microorganisms exhibiting diverse morphologies provided one of the earliest glimpses into the remarkable complexity of the oral microbial ecosystem (71-73).
Positioned at the forefront of the alimentary tract, the oral cavity sustains a finely tuned microbial equilibrium that underpins both oral and systemic physiological integrity. The oral cavity comprises multiple distinct ecological niches, including the teeth, buccal mucosa, soft and hard palates, and tongue, which together form a highly complex microenvironment. This fosters the coexistence of diverse microbial consortia, collectively referred to as the oral microbiome (74). Oral microbiome dysbiosis is increasingly recognized as a contributing factor in the pathogenesis of a broad spectrum of oral and systemic disorders. Perturbations in this homeostasis, collectively referred to as oral dysbiosis, have been implicated as pivotal contributors to a spectrum of pathological processes. Among these, the intricate and multifaceted interplay between oral microbial dysregulation and oral carcinogenesis has garnered substantial scholarly interest. Notably, malignant transformation within the oral epithelium can actively reshape the resident microbiota, thereby creating a niche increasingly conducive to tumor persistence and progression (75-80).
The term microbiome refers to the collective assemblage of symbiotic, commensal, and pathogenic microorganisms inhabiting a defined ecological niche (81). The oral cavity harbors a vast and diverse array of microorganisms and remains in continuous interaction with the external environment, rendering it particularly susceptible to environmental influences (82). The oral microbiome is a complex consortium of bacteria, fungi, viruses, archaea, and protozoa that collectively contribute to the establishment and maintenance of its normal microbial community (81). Bacteria represent the principal constituents, assembling into habitat-specific microbial consortia across the various niches of the oral cavity.
Investigations into the oral microbiome have identified a remarkable diversity comprising more than 700 bacterial species, which are taxonomically distributed across seven principal phyla: Bacteroidota (Bacteroidetes), Actinomycota (formerly Actinobacteria), Fusobacteriota (Fusobacteria), Bacillota (Firmicutes), Pseudomonadota (Proteobacteria), Spirochaetota (Spirochaetes), and Saccharibacteria. Despite this diversity, most species are derived from only a few dozen genera (83-85). The oral microbiome is characterized by pronounced spatial and temporal variability, exhibiting rapid shifts in both community composition and functional activity that evolve in parallel with host development. These complex, non-equilibrium dynamics arise from a confluence of factors, including dietary components and alterations in local pH, as well as interbacterial interactions that confer novel functional attributes on microbial strains (86). The predominant bacterial taxa that constitute the core of the oral microbiome are conserved primarily across individuals, reflecting a stable and shared microbial framework despite inter-individual variability in less abundant species. The predominant bacterial genera characterizing a healthy oral cavity are summarized in Table 3.
Table 3. Taxonomic profile of the major bacterial genera in the healthy oral microbiome.
| Cocci | Rods | |
|---|---|---|
| Gram positive | Abiotrophia, Peptostreptococcus, Streptococcus, Stomatococcus. | Actinomyces, Bifidobacterium, Corynebacterium, Eubacterium, Lactobacillus, Propionibacterium, Pseudoramibacter, Rothia |
| Gram negative | Moraxella, Neisseria, Veillonella | Campylobacter, Capnocytophaga, Desulfobacter, Desulfovibrio, Eikenella, Fusobacterium, Hemophilus, Leptotrichia, Prevotella, Selemonas, Simonsiella, Treponema, Wolinella. |
Beyond bacterial populations, the oral microbiota also comprises diverse microeukaryotes, such as fungi, amoebae, and flagellates, as well as archaeal species and a broad spectrum of viruses (87). In most individuals, the oral mycobiome is composed primarily of fungal species belonging to the genera Candida and Malassezia (88-93). The oral microbiota exerts a pivotal influence on oral health, as three of the most common oral pathologies, dental caries, periodontal disease, and oral cancer, are primarily driven by microbial etiologies. Several extensively characterized periodontal microbiomes have been identified as central to elucidating the mechanistic links between oral microbial dysbiosis and oncogenesis (94). Oral squamous cell carcinoma (OSCC) represents the predominant malignancy of the head and neck region, comprising nearly 2% of all cancer diagnoses worldwide (95). While traditionally linked to lifestyle risk factors such as tobacco use and excessive alcohol consumption, emerging evidence implicates specific constituents of the oral microbiome in OSCC pathogenesis. Among these, Porphyromonas gingivalis has garnered particular attention, given its well-documented association with both the initiation and progression of neoplastic transformation within the oral cavity (96). In a comparative analysis of microbial communities within OSCC lesions and contralateral healthy tissues from 50 patients, Zhang et al. reported a significant enrichment of Porphyromonas species in tumor-associated samples (97). This observation is consistent with findings by Katz et al., who documented elevated levels of P. gingivalis in gingival specimens from patients with OSCC compared with healthy controls (98). Together, these studies underscore that microbial communities differ markedly between malignant and adjacent healthy oral tissues, with tumor sites harboring a greater abundance of pathogenic taxa. A systematic review and meta-analysis by Sayehmiri et al. further confirmed this association (99), revealing that colonization by P. gingivalis was associated with an increased risk of oral cancer (odds ratio, 1.36), with gingival cancers accounting for most cases. Experimental evidence also supports these observations: in a murine model, Wen et al. demonstrated that P. gingivalis infection promoted tumor multiplicity and growth and accelerated malignant progression (100). Beyond P. gingivalis, Rai et al. recently demonstrated that Porphyromonas endodontalis was also enriched in the salivary microbiota of patients with OSCC, suggesting that multiple Porphyromonas species may contribute to oral tumorigenesis (101).
As the lungs and oral cavity are connected, the composition and dynamics of the oral microbiome are closely linked to those of the lung microbiome. Migration of oral bacteria into the lower respiratory tract represents a key pathogenic mechanism underlying aspiration pneumonia (102). Likewise, their colonization of the gastrointestinal tract, often intensified by dysregulated gastric or bile acid secretion in systemic disorders such as cirrhosis, has been associated with the development of inflammatory bowel disease and colorectal cancer (103-107).
In addition, the carriage of specific oral microbiota has been linked to an increased susceptibility to pancreatic cancer (PC) (108-111). Multiple studies have reported significant associations between Porphyromonas gingivalis and PC, while Mitsuhashi et al. demonstrated that the intratumoral presence of Fusobacterium nucleatum correlates with poorer clinical outcomes (112). Beyond these organisms, Fan et al. further identified Aggregatibacter actinomycetemcomitans and Alloprevotella as associated with an increased risk of PC development (108. Moreover, Wei et al. reported that colonization by Leptotrichia and Streptococcus species is also associated with an elevated risk of pancreatic cancer (113). In one of the earliest investigations into the association between the oral microbiota and PC, Farrell et al. identified an enrichment of Granulicatella adiacens in patients with PC. Additionally, their analysis revealed differential abundances of Neisseria elongata and Streptococcus mitis between affected individuals and healthy controls (114). It remains uncertain whether these microbes are true oncogenic drivers or secondary colonizers of the tumor niche.
2.3. Gastric cancer
Gastric cancer (GC) ranks among the most prevalent malignancies and remains a leading contributor to global cancer-related mortality (115, 116). GC was initially categorized based on histopathological and anatomical criteria; however, these conventional classifications proved inadequate for guiding therapeutic decision-making and yielded only a slight improvement in patient outcomes. More recently, clinical and molecular profiling has emerged as a more reliable framework for stratifying patients and tailoring treatment strategies. Genomic approaches have been particularly instrumental in delineating molecular subtypes of GC. In 2011, Tan et al. proposed two distinct genomic variants-the genomic intestinal (G-INT) and genomic diffuse (G-DIF) subtypes, characterized by unique histological features, gene expression signatures, biological pathways, and prognostic implications. These molecular subtypes partially overlap with Lauren’s traditional classification, reflecting the profound clinical and biological heterogeneity inherent to GC, mainly attributable to the diverse molecular landscapes of malignant cells (117). GC is rarely diagnosed at an early stage, which substantially restricts therapeutic options. Its biological complexity continues to obscure a comprehensive understanding of disease mechanisms, thereby posing significant challenges to effective management and eradication. The development of GC represents the culmination of a multifaceted interaction among host genetic susceptibilities, environmental exposures such as tobacco use, alcohol consumption, high dietary salt and meat intake, and insufficient consumption of fruits and vegetables, and microbial influences, most notably Helicobacter pylori infection and alterations within the gastric microbiome (118-120). A defining feature of GC lies in its intricate relationship with the resident microbial ecosystem of the stomach. While Helicobacter pylori has long been established as the primary initiator of gastric carcinogenesis, emerging evidence highlights the broader contribution of diverse microbial inhabitants of the gastric mucosa to disease progression (121). Perturbations in the gastric microbiota appear to orchestrate key events across the carcinogenic continuum, spanning the transition from premalignant alterations to the establishment of invasive gastric cancer (122-125).
The human gastrointestinal tract harbors a highly diverse microbial ecosystem, collectively referred to as the gut microbiome. This community is primarily composed of four dominant bacterial phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. Among these, Firmicutes, including genera such as Clostridium, Ruminococcus, Eubacterium, Dorea, Peptostreptococcus, and Lactobacillus, are the most prevalent, accounting for approximately 30.6%-83% of the total microbiota. Bacteroidetes, primarily represented by Bacteroides, constitute 8-48%, whereas Actinobacteria, dominated by Bifidobacterium, contribute 0.7-16.7%. Proteobacteria, including members of the Enterobacteriaceae, make up a variable fraction ranging from 0.1-26.6% (126). Alterations in microbial composition can impair the equilibrium between the gut microbiota and the host immune system, thereby predisposing the intestinal environment to chronic inflammation and subsequent oncogenic transformation.
Extensive research has established Helicobacter pylori as a central factor in gastric cancer (GC) pathogenesis. Its discovery not only overturned the long-standing belief that the acidic stomach is sterile but also marked the identification of the only bacterial species thus far classified as a class I carcinogen. Although spiral-shaped microorganisms in the stomach had been observed earlier, it was not until 1982 that Warren and Marshall conclusively linked bacterial infection to chronic gastritis and successfully isolated the causative organism (127, 128). The gastric environment exhibits a steep pH gradient, ranging from 1 to 2 within the gastric lumen to 6 to 7 along the mucosal surface, with the latter providing a more favorable niche for microbial colonization (129, 130). Bacteria typically enter the stomach from the upper digestive or respiratory tracts. Among these, Helicobacter pylori has uniquely adapted to survive in the acidic milieu of the stomach and is recognized as a key etiological agent of noncardiac gastric adenocarcinomas. This Gram-negative, spiral-shaped, flagellated member of the phylum Proteobacteria exhibits urease, catalase, and oxidase activities, which facilitate its persistence in the gastric niche (131, 132). H. pylori is characterized by high motility conferred by a unipolar bundle of sheathed flagella (133). Clinically, H. pylori infection is strongly implicated in the pathogenesis of chronic gastritis, atrophic gastritis, mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric adenocarcinoma (134).
H. pylori promotes gastric carcinogenesis by inducing direct genotoxic stress, primarily through the conversion of nitrogenous compounds in gastric fluid into carcinogenic N-nitroso compounds (NOCs) and reactive nitrogen intermediates, while simultaneously fostering a chronic pro-inflammatory microenvironment within the gastric mucosa (135). The oncogenic potential of H. pylori is largely attributed to two major virulence determinants: cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA), which perturb host cell functions and activate oncogenic signaling pathways (136, 137). CagA, a strain-specific effector protein delivered into host epithelial cells via the H. pylori type IV secretion system, functions as a classical oncogene. Its activity contributes to chronic gastritis, peptic ulcer disease, MALT lymphoma, and gastric carcinoma. Mechanistically, CagA disrupts epithelial homeostasis by suppressing apoptotic pathways and inducing morphological abnormalities, such as cell scattering, elongation, and loss of polarity (137). VacA represents another major H. pylori virulence determinant, functioning as a multifunctional exotoxin that induces diverse pathological effects in host cells, including vacuolization, apoptosis, and necrosis. Beyond these cytotoxic properties, VacA integrates into host cell membranes, where it behaves as an anion-selective channel. Through this channel activity, VacA facilitates the efflux of bicarbonate and organic anions into the cytoplasm, which enhances H. pylori colonization and persistence within the gastric niche (138). H. pylori infection elicits chronic inflammation within the gastric mucosa, a recognized antecedent of neoplastic transformation (139). H. pylori also induces inflammatory responses in gastric epithelial cells primarily through activation of NF-κB, which drives the secretion of proinflammatory cytokines, including interleukin (IL)-1β, IL-2, IL-6, IL-7, IL-8, IL-10, interferon-γ (IFN-γ), and tumor necrosis factor-α (TNF-α). In addition, H. pylori promotes inflammation by upregulating cyclooxygenase-2 (COX-2), thereby increasing prostaglandin E2 (PGE2) production (139).
Accumulating evidence indicates that, beyond H. pylori, other constituents of the gastric microbiota play critical roles in driving malignant transformation. For example, fungi and viruses may also contribute to the multifactorial processes underlying gastric carcinogenesis. A study by Zhong M et al. identified a GC-associated mycobiome imbalance characterized by disrupted fungal composition and ecology, highlighting Candida albicans as a potential fungal biomarker for gastric cancer. In GC samples, the relative abundance of C. albicans, Fusicolla acetilerea, Arcopilus aureus, and Fusicolla aquaeductuum was markedly elevated, whereas Candida glabrata, Aspergillus montevidensis, Saitozyma podzolica, and Penicillium arenicola were significantly reduced. Moreover, C. albicans may contribute to gastric carcinogenesis by reducing fungal richness and diversity in the stomach, thereby facilitating disease progression (13). Fungal dysbiosis in the stomach has been shown to activate inflammatory pathways, including cytokine and chemokine signaling. In the context of impaired immune responses, particularly in patients with advanced-stage GC, this imbalance increases susceptibility to opportunistic fungal infections. However, whether the enrichment of specific fungi in GC is a driving factor in immune dysregulation or merely a consequence of tumor-associated changes remains unresolved, and their potential roles as oncogenic pathogens warrant further investigation (140, 141). Epstein-Barr virus (EBV) accounts for approximately 7-9% of global gastric cancer cases annually and promotes carcinogenesis through extensive genomic and epigenomic alterations (142). EBV-driven amplification and overexpression of programmed death ligand 1 (PD-L1) enable tumor cells to evade T cell-mediated immunity, while latency-associated products, including EBV nuclear antigen 1, latent membrane protein 2A, and viral microRNAs, further contribute to oncogenesis by inducing epigenetic dysregulation and aberrant mRNA transcription (143, 144). Although other viruses, such as human papillomavirus, human herpesvirus, and hepatitis viruses, have been implicated in GC, no definitive causal role has been established. Overall, the gastric virome remains poorly characterized and warrants further investigation (145).
2.4. Pancreatic cancer
Pancreatic cancer (PC) represents one of the most lethal and aggressive malignancies, with a rising incidence globally. In the United States, the current 5-year overall survival rate remains dismal at only 10.8%. Broadly, pancreatic cancers are classified into two major categories: pancreatic ductal adenocarcinoma (PDAC) constitutes over 90% of all pancreatic malignancies, representing the predominant histological subtype (146), and pancreatic neuroendocrine tumors (PanNETs), a less common but biologically distinct entity (147). PDAC is a highly aggressive malignancy characterized by an exceptionally poor prognosis. This unfavorable outcome is primarily attributed to its frequent diagnosis at advanced, often unresectable stages, coupled with a high degree of intrinsic and acquired resistance to conventional therapies. Surgical resection remains the sole potentially curative treatment for pancreatic ductal adenocarcinoma; however, only approximately 20% of patients present with tumors amenable to resection at the time of diagnosis (148). Despite intensive research, the molecular mechanisms driving PDAC oncogenesis and its profound treatment refractoriness remain incompletely understood (149). The development of pancreatic cancer is driven by a multifactorial interplay of influences, including genetic alterations, lifestyle factors, particularly smoking and high-fat diets; dysbiosis of the gut microbiota; and comorbid conditions such as obesity, type 2 diabetes, and chronic pancreatitis, among others (150-154). Given the poor long-term outcomes of PDAC and the limited efficacy of current systemic therapies, there is an urgent need to develop novel therapeutic approaches and supportive strategies that aim to improve patients' quality of life. Increasing attention has recently been directed toward the relationship between pancreatic cancer and the microbiome. In PDAC, dysbiosis involving bacterial, fungal, and viral communities has been consistently reported (155). Thus, modulation of the gut microbiome and restoration of its ecological balance may represent a promising avenue for therapeutic intervention.
Historically, the pancreas was regarded as a sterile organ, much like the lung. However, recent advances in sequencing technologies have revealed that pancreatic tissue harbors its own distinct microbiota (156). In a seminal study, Pushalkar et al. used 16S rRNA gene sequencing and demonstrated that Proteobacteria, Bacteroidetes, and Firmicutes were significantly enriched in pancreatic cancer tissue compared with normal pancreatic tissue (157). Accumulating evidence now suggests that the intratumoral microbiome plays a crucial role in the initiation, progression, and prognosis of pancreatic cancer (154, 157-161). These effects are mediated mainly by microbial modulation of host immune responses and by alterations in drug metabolism, thereby influencing both tumor biology and therapeutic outcomes. Multiple epidemiological and mechanistic studies have highlighted the contribution of periodontal disease and tooth loss to pancreatic carcinogenesis. A comprehensive meta-analysis reported a strong association between periodontal pathologies, particularly the presence of Porphyromonas gingivalis, and increased risk of pancreatic cancer (162). Furthermore, several investigations have explored the relationship between specific oral pathogens, including P. gingivalis, Fusobacterium spp., Neisseria elongata, and Streptococcus mitis, and the development of PDAC. Among these, P. gingivalis consistently shows the strongest positive correlation with PDAC susceptibility, suggesting a potential role as a microbial risk factor in pancreatic tumorigenesis (163, 164). Emerging evidence indicates that fungal and viral infections may contribute to the pathogenesis of pancreatic cancer (PC).
A study by Aykut et al. demonstrated that the intrapancreatic mycobiome, particularly enriched with Malassezia spp., is closely associated with the development and progression of PDAC. The fungal composition of tumor tissue was distinct from that of the gut or normal pancreatic tissue. Notably, experimental ablation of the mycobiome suppressed tumor growth in both slowly progressive and invasive murine PDAC models, whereas repopulation with Malassezia spp. accelerated oncogenesis. Mechanistic investigations revealed that ligation of mannose-binding lectin (MBL), which recognizes glycans on the fungal cell wall and activates the complement cascade, is essential for this tumor-promoting effect (154). Additional evidence links Candida infection with pancreatic cancer risk. A prospective cohort study in Sweden identified an association between oral Candida colonization and an increased incidence of PC (165). Mechanistically, Candida may drive tumorigenesis by inducing chronic inflammation and promoting the expansion of myeloid-derived suppressor cells (MDSCs), thereby fostering an immunosuppressive tumor microenvironment (150). Viruses have also been implicated in pancreatic carcinogenesis. Several studies have reported associations between chronic pancreatitis and hepatitis B virus (HBV) infection, while a meta-analysis by Arafa et al. demonstrated that hepatitis C virus (HCV) infection significantly increases the risk of PC (166-168). These findings suggest that chronic viral infections, through persistent inflammation and pancreatic injury, may serve as cofactors in pancreatic tumorigenesis. Collectively, these studies highlight the potential oncogenic roles of fungi and viruses in PC, warranting further mechanistic and clinical investigations.
2.5. Colorectal cancer
Colorectal cancer (CRC) is the most prevalent malignancy of the digestive tract and represents a major global health burden. Accounting for approximately 10% of all cancer diagnoses, CRC is currently the third most common cancer worldwide and ranks among the leading causes of cancer-related mortality. Recent estimates indicate nearly 700,000 deaths annually, underscoring its persistently high morbidity and mortality rates. While CRC was considered relatively uncommon several decades ago, its incidence has risen sharply, making it one of the most lethal cancers globally (95, 169). The global burden of colorectal cancer is exacerbated not only by demographic transitions, such as population ageing, and the prevalence of Westernized dietary habits, but also by modifiable lifestyle determinants, including obesity, sedentary behavior, and tobacco use. Collectively, these factors amplify disease incidence and mortality, rendering colorectal cancer a formidable challenge to healthcare systems across the world (170).
The gut microbiome has increasingly been recognized as a pivotal determinant in human health and disease, with mounting evidence highlighting its relevance in CRC. Numerous investigations have demonstrated that alterations in microbial composition, shaped by dietary patterns and environmental exposures, can promote CRC development through mechanisms involving chronic inflammation, bioactive microbial metabolites, and pathogenic virulence factors. Beyond tumor initiation, dysbiosis of the gut microbiota also exerts a profound influence on CRC progression and trajectory (170-172). Fusobacterium nucleatum has emerged as one of the most extensively studied bacterial taxa implicated in colorectal carcinogenesis. Metagenomic profiling consistently associates Fusobacterium spp. with CRC, although the precise nature of this relationship, causal or correlative, remains unresolved. Castellarin et al. reported a nearly 400-fold increase in F. nucleatum transcript levels in CRC tissues relative to adjacent normal mucosa, underscoring its enrichment in the tumor microenvironment. In an (APC)+/− mouse model, F. nucleatum promoted neoplastic progression by creating a pro-inflammatory milieu within intestinal epithelial cells and facilitating the recruitment of tumor-infiltrating immune cells (173, 174). Elevated IL-17a expression has also been observed in CRC patients with abundant F. nucleatum, suggesting a role in inflammation-driven tumorigenesis. Mechanistically, this strain exhibits strong mucosal adherence and produces Fusobacterium adhesin A (FadA), a virulence factor that binds to E-cadherin and activates β-catenin signaling, thereby driving oncogenic pathways (175, 176). Notably, F. nucleatum has been associated with consensus molecular subtype 1 (CMS1) CRC, characterized by microsatellite instability and immune pathway upregulation (177, 178). More recently, studies of metastatic CRC demonstrated that nearly identical strains of Fusobacterium persist in both primary tumors and distant metastases, highlighting its potential role as a stable component of the tumor microenvironment and a facilitator of disease dissemination (179).
Enterotoxigenic Bacteroides fragilis (ETBF), a strain that produces B. fragilis toxin (BFT), is implicated not only in diarrheal disease and inflammatory bowel disease but also in colorectal tumorigenesis (180, 181). Mechanistically, ETBF promotes tumor development by activating STAT3 signaling and driving a Th17-mediated inflammatory response (182). Colonization with BFT+ B. fragilis also promotes the accumulation of regulatory T cells, thereby amplifying IL-17-driven procarcinogenic inflammation (183). In epithelial cells, BFT induces cleavage of E-cadherin, thereby increasing paracellular permeability and activating β-catenin signaling, ultimately enhancing proliferative capacity (184). Beyond direct host signaling, BFT+ B. fragilis perturbs the gut microbial ecosystem by fostering dysbiosis, encouraging the outgrowth of other procarcinogenic taxa, impairing mucosal immune defenses, disrupting epithelial barrier integrity, and promoting mucin degradation (183-186).
Pathogenic Escherichia coli harboring the pks genomic island represents another gut-associated bacterium that is strongly enriched in CRC tissues and is functionally linked to tumor promotion in preclinical models. Strains carrying the pks island secrete a family of heat-labile cytolethal distending toxins that colonize the intestinal mucosa, elicit inflammation, and increase the host's mutational burden (187). Moreover, pks⁺ E. coli encodes the genotoxic polyketide-peptide hybrid colibactin, which, upon delivery to eukaryotic cells, induces DNA double-strand breaks, disrupts the cell cycle, and generates chromosomal abnormalities. These combined mutagenic and pro-inflammatory effects establish pks⁺ E. coli as a potent microbial driver of colorectal tumorigenesis (20, 188).
Among tumor-associated microbes, Fusobacterium nucleatum and pks⁺ Escherichia coli are among the most intensively studied species. Both contribute to colorectal tumorigenesis through intertwined mechanisms of inflammation, genotoxicity, and immune modulation (187). F. nucleatum promotes chronic inflammation by activating the NF-κB pathway and inducing cytokines such as IL-6 and TNF-α. Its adhesin, FadA, facilitates β-catenin signaling, thereby enhancing epithelial proliferation, whereas its Fap2 protein binds to TIGIT on T cells and NK cells, leading to promoting immune evasion. Conversely, pks⁺ E. coli produces colibactin, a genotoxin that causes DNA double-strand breaks and generates a characteristic mutational signature identified in human colorectal tumors (187). Despite compelling mechanistic data, the exact oncogenic role of these microbes remains a matter of controversy. Some studies suggest that F. nucleatum colonizes pre-existing lesions rather than initiating cancer, whereas others show that its depletion reduces tumor burden in animal models. Similarly, colibactin’s genotoxicity is context-dependent, varying with host DNA-repair capacity and microbial abundance. Furthermore, both bacteria can reshape the tumor microenvironment, either by amplifying inflammation or promoting immunosuppression, depending on tumor stage and host immunity. Integrating these findings suggests that F. nucleatum and pks⁺ E. coli act not as single “drivers,” but as dynamic modulators within the complex microbial ecosystem, influencing tumor evolution.
2.6. Hepatocellular carcinoma
Globally, hepatocellular carcinoma (HCC) is recognized as a highly prevalent malignancy and a foremost cause of cancer mortality (189, 190). Major etiological factors include persistent infection with the hepatitis B virus (HBV) or the hepatitis C virus (HCV), alcoholic liver disease, and non-alcoholic fatty liver disease (NAFLD). These conditions drive progressive hepatic injury and fibrogenesis, culminating in cirrhosis, which constitutes the principal precursor state for HCC development (191).
Gut microbiome dysbiosis is a characteristic feature of patients with HCC, typically characterized by an expansion of pathogenic taxa and a depletion of commensal, health-promoting bacteria. In a study by Zhang et al., hepatocellular carcinoma patients stratified by the Barcelona Clinic Liver Cancer staging system exhibited progressive alterations in gut microbiota composition, characterized by increased abundances of Enterococcus and Enterobacteriaceae and concomitant reductions in Actinobacteria and Bifidobacterium, with advancing disease severity (192). In a study conducted by Zheng et al., comparative analysis across cohorts of patients with hepatitis, cirrhosis, cirrhosis-associated HCC, non-cirrhosis-related HCC, and healthy controls revealed that HCC patients exhibited a significant enrichment of Bacteroidetes and Fusobacteria, along with increased gut microbial diversity relative to the other groups (193). Wang et al. provided compelling evidence for a causal role of gut dysbiosis in hepatocarcinogenesis. Using fecal microbiota transplantation (FMT) from patients with HCC and healthy controls into germ-free and specific-pathogen-free (SPF) mice, they demonstrated that reconstitution with HCC-associated microbiota induced spontaneous liver inflammation, fibrosis, and dysplasia, and accelerated chemically induced HCC. Mechanistically, HCC-derived microbiota disrupts intestinal barrier integrity, facilitating the translocation of viable pathogenic bacteria into the liver and triggering pro-inflammatory cascades that sustain tumorigenesis. Notably, both murine and human livers showed enrichment of Klebsiella pneumoniae, and monocolonization with this species recapitulated the tumor-promoting effects of HCC-FMT, thereby establishing K. pneumoniae as a key oncogenic driver in HCC (194). Moreover, the dynamic crosstalk between bile acids (BAs) and the gut microbiota has emerged as a pivotal determinant in the initiation and progression of HCC. Under physiological conditions, BA metabolism is tightly orchestrated through bidirectional interactions between host and microbial communities, whereby gut microorganisms modulate BA composition and BAs act as signaling molecules to preserve hepatic and intestinal homeostasis. However, dysbiosis of the gut microbiota in chronic liver disease and malignant transformation perturbs BA equilibrium, thereby fostering hepatic inflammation and fibrogenesis and ultimately driving hepatocarcinogenesis (195).
The liver maintains a tightly interconnected bidirectional communication with the gut microbiota, commonly referred to as the gut-liver axis. Microbial communities and their metabolites exert a profound influence on hepatic homeostasis, while the disruption of this equilibrium, termed dysbiosis, has been implicated in the pathogenesis of diverse liver disorders (196, 197). Mechanistically, microbial dysbiosis promotes hepatic injury and inflammation by compromising intestinal barrier integrity, thereby facilitating bacterial translocation and exposure of the liver to microbial products and pathogen-associated molecular patterns. For instance, studies have demonstrated that elevated systemic levels of zonula occludens-1 (ZO-1), a tight junction protein, correlate with increased intestinal permeability, heightened inflammatory responses, and greater disease severity in patients with HCC (198, 199). Disruption of intestinal barrier integrity permits the translocation of microbial products, most notably lipopolysaccharide (LPS), thereby delivering potent pro-inflammatory cues from the gut lumen directly into the hepatic milieu (197, 198, 200, 201). LPS engages toll-like receptor 4 (TLR4), triggering the downstream activation of the NF-κB signaling cascade and the consequent secretion of pro-inflammatory cytokines. Under conditions of dysbiosis, bacterial overgrowth exacerbates the TLR4-NF-κB-mediated inflammatory axis, thereby fostering persistent intestinal inflammation and driving hepatocarcinogenesis (202, 203).
2.7. Breast cancer
Breast cancer (BC) is the most frequently diagnosed malignancy among women and, despite considerable advances in diagnostic approaches and therapeutic strategies, it continues to rank as a leading cause of cancer-related mortality worldwide (204). Breast cancer represents a heterogeneous malignancy comprising distinct subtypes with unique epidemiological features (205). Globally, it accounts for approximately one-third of all cancers diagnosed in women, with mortality contributing to nearly 15% of cases (206, 207). A multifactorial interplay of genetic predisposition, environmental exposures, and lifestyle determinants shapes the worldwide distribution of breast cancer. While incidence rates are typically higher in high-income countries, mortality is comparatively lower due to the availability of early detection programs and more effective therapeutic interventions, in contrast to resource-limited settings (208). In recent years, the microbiome has emerged as a novel factor potentially linked to BC. As a fundamental regulator of human health and homeostasis, the microbiome exerts broad effects on biological, hormonal, and metabolic pathways. Through these mechanisms, it may influence tumor initiation, proliferation, and genomic instability in host cells, whereas in other contexts it can promote apoptosis and tumor suppression (209, 210).
Complementary work by Smith et al. revealed that the breast tissue microbiome exhibits variability across racial groups, tumor stages, and molecular subtypes, underscoring its potential role in shaping disease heterogeneity (211). In a large cohort study, Thompson et al. demonstrated a significant association between the breast microbiota and host gene expression, identifying bacterial taxa that correlated with molecular programs governing epithelial-mesenchymal transition (EMT) and cellular proliferation. More recent investigations have further highlighted the functional importance of intratumoral microbiota, demonstrating that these microorganisms facilitate breast cancer metastasis by enhancing cellular resistance to fluid shear stress through actin cytoskeletal remodeling, thereby promoting tumor cell survival and dissemination (212). Collectively, these findings underscore that intratumoral microorganisms are not merely incidental but are pervasive within breast cancer tissues, where they may actively influence disease initiation, progression, and clinical outcome.
Multiple studies have demonstrated that the microbial composition of mammary gland tissue undergoes distinct alterations between malignant and non-malignant states, as well as across different tumor stages (213, 214). Xuan et al. identified Sphingomonas yanoikuyae as a commensal organism in normal breast tissue, which was markedly depleted in tumor samples. At the same time, Methylobacterium radiotolerans emerged as the most significantly enriched bacterium within tumor tissue (215). In an Asian breast cancer cohort, tumor tissues were found to harbor increased abundances of Propionicimonas, Micrococcaceae, Caulobacteraceae, Rhodobacteraceae, Nocardioidaceae, and Methylobacteriaceae, accompanied by a reduction in Bacteroidaceae (216). Notably, disease progression was associated with a concomitant enrichment of the genus Agrococcus. Furthermore, advanced malignancy was associated with increased prevalence of Fusobacterium, Atopobium, Gluconacetobacter, Hydrogenophaga, and Lactobacillus, highlighting a progressive remodeling of the breast tumor microbiome in relation to oncogenesis (217).
Endogenous estrogen plays a pivotal role in the pathogenesis of breast cancer, particularly in the postmenopausal setting, where approximately 70% of tumors are classified as estrogen receptor positive. Before menopause, the ovaries serve as the primary site of estrogen biosynthesis, and circulating estrogens exert systemic endocrine effects on various target tissues, including the skeletal, neural, and immune systems (218). Following hepatic metabolism, estrogens and their derivatives undergo conjugation via glucuronidation and sulfonation, processes that facilitate their excretion through bile. Although a substantial fraction of these conjugated metabolites is eliminated in urine and feces, a considerable proportion undergoes enterohepatic recirculation. This is mediated by gut microbes that express β-glucuronidase activity, which hydrolyze conjugated estrogens to their bioactive forms, thereby facilitating reabsorption into the systemic circulation. Moreover, intestinal microorganisms can generate estrogenic compounds or structural mimics from dietary substrates, further influencing host estrogen homeostasis (218). β-glucuronidase is a central enzymatic component of the estrobolome, deconjugating estrogens and thereby restoring their bioactive forms for reabsorption into the systemic circulation.
Recent work refines the “estrobolome” concept, the ensemble of gut-microbial genes (notably β-glucuronidases) that deconjugate hepatically conjugated estrogens excreted in bile, thereby enabling enterohepatic reabsorption and altering systemic estrogen exposure relevant to ER⁺ disease. Contemporary reviews map estrobolome enzymatic targets/taxa and propose standardized measurement panels to align microbiome endpoints with breast cancer risk and therapy studies (219, 220). Large 2024-2025 syntheses and narrative updates collectively report links between gut/breast microbial signatures and tumor risk, subtype, and treatment response, yet emphasize heterogeneity across cohorts and the current inability to meta-analyze genera consistently associated with outcomes (Table 4).
Table 4. Recent updates (2024-2025) for breast and prostate microbiome research.
| Domain | Key 2024-2025 insights | Implications |
|---|---|---|
| Breast - Estrobolome and estrogen | Estrobolome targets mapped; β-glucuronidase-mediated deconjugation drives enterohepatic estrogen recycling; tissue and multi-kingdom signals revisited with stricter controls. | Align microbiome endpoints with ER⁺ risk/therapy; prioritize standardized assays for Estrobolome activity. |
| Breast - Evidence synthesis | 2025 systematic review: 48 studies; heterogeneity precludes genus-level meta-analysis; stool and tissue datasets dominate; need for harmonized pipelines. | Standardize sampling/bioinformatics; design longitudinal/interventional studies. |
| Prostate - Urinary microbiome | Reviews emphasize urinary/tissue microbiomes as non-invasive biomarkers, methodology standardization outstanding. | Develop validated urine microbiome pipelines for screening and risk stratification. |
| Prostate - Gut - prostate axis and hormones | Gut microbes can influence androgen pathways and ADT response; lower α-diversity correlates with tumor burden; cross-species models support hormonal crosstalk. | Integrate microbiome-hormone multi-omics; test microbial modulation alongside hormonal therapy. |
2.8. Prostate cancer
Prostate cancer (PCa) represents the second most frequently diagnosed malignancy in men worldwide. It remains a leading cause of cancer-related mortality, accounting for approximately 1.6 million new cases and 366,000 deaths each year (221, 222). Epidemiological and observational studies provide compelling evidence that unhealthy dietary patterns, excessive alcohol intake, and tobacco use are strongly associated with an elevated risk of chronic non-communicable diseases, including various malignancies (223). Nonetheless, the precise contribution of these lifestyle factors to prostate cancer (PCa) pathogenesis remains inconclusive. Emerging evidence has increasingly underscored the role of the human microbiota, particularly the gut microbiota (GM), in shaping disease susceptibility and progression. As a result, microbial communities residing in the gut have garnered considerable attention for their potential influence on host physiology and their implications in PCa development (224, 225). In 2018, Liss et al. analyzed the gut microbiota in 133 American men undergoing prostate biopsy and, for the first time, demonstrated a potential association between the gut microbiome and prostate cancer (226). In a separate study, Bacteroides massiliensis was found to be more prevalent in the gut microbiota of Caucasian men with prostate cancer compared to those with benign prostatic hyperplasia (BPH). In contrast, Faecalibacterium prausnitzii and Eubacterium rectalis were observed at reduced levels (227).
The gut microbiota has recently been conceptualized as an androgen-producing “organ.” Emerging evidence indicates that microbial metabolites can influence prostate cancer growth and progression, supporting the existence of a “gut-prostate axis” (228). Androgens are central drivers of prostate cancer, exerting their effects through binding to the androgen receptor in malignant prostate cells. While testosterone is primarily synthesized in the testes and dehydroepiandrosterone (DHEA) in the adrenal glands (229), several studies suggest that the gut microbiota also contributes to androgen biosynthesis and regulation, thereby potentially shaping the hormonal milieu that governs prostate cancer development. Matsushita M et al. collected rectal swab samples from Japanese male subjects who were clinically suspected of having prostate cancer and underwent prostate biopsy. To minimize confounding factors, individuals with positive biopsy results were excluded, ensuring that only patients without prostate cancer were included in the analysis. In this cohort of elderly men, we investigated the association between gut microbiota composition and circulating testosterone levels. Microbial community diversity, assessed using both α- and β-diversity indices, did not differ significantly by testosterone status. However, taxonomic profiling revealed that specific genera within the phylum Firmicutes were more prevalent in subjects with higher total testosterone (TT) levels.
Notably, beyond gut dysbiosis, recent studies have profiled the urinary and prostate-tissue microbiomes as potential noninvasive biomarkers for early detection and risk stratification (230). Reviews summarize that distinct urinary/gut consortia correlate with incidence, grade, and treatment outcomes; however, causality remains unresolved, and standardization is needed for specimen collection, sequencing, and batch control (Table 3). Mechanistically, the gut-prostate axis encompasses microbial effects on androgen metabolism (e.g., steroidogenic pathways and 5α-reductase activity), immune tone, and tumor energetics, features implicated in castration resistance and response to androgen-deprivation therapy (ADT). Longitudinal translational work reports that reduced fecal α-diversity correlates with tumor burden in hormonotherapy-naïve PCa, and that microbial community shifts may modulate hormonal treatment responses. Parallel reviews call for integrated multi-omics and prospective designs to resolve directionality and identify therapeutic leverage points.
2.9. Gynecological cancers
Cervical cancer ranks as the fourth most prevalent malignancy among women, accounting for an estimated 342,000 deaths in 2020. More than 95% of cases are attributable to persistent infection with human papillomavirus (HPV), a pathogen with exceptionally high prevalence, as over 70% of sexually active women are estimated to acquire infection during their lifetime (231, 232). Histologically, cervical cancer is predominantly classified into two subtypes: squamous-cell carcinoma (SCC), which constitutes the majority, and adenocarcinoma (ADC) (233). Increasing evidence indicates that the vaginal microbiota exerts a significant influence on both cervical carcinogenesis and the persistence or clearance of HPV. Brotman, R. M., et al. have reported that Lactobacillus gasseri abundance correlates with viral clearance, whereas Atopobium spp. is strongly associated with HPV persistence (234). Moreover, the vaginal microbiota of women with cervical intraepithelial neoplasia or cervical cancer is characterized by marked depletion of Lactobacillus spp. compared to healthy counterparts, alongside enrichment of taxa frequently linked to bacterial vaginosis, including Gardnerella, Megasphaera, Prevotella, Peptostreptococcus, Streptococcus, Sneathia sanguinegens, and Atopobium. These microbial shifts suggest a dysbiotic microenvironment that may facilitate viral persistence and malignant transformation (235).
Endometrial cancer (EC), arising from the epithelial lining of the uterine cavity, represents a malignancy with steadily increasing incidence and associated mortality worldwide. Traditionally, EC has been stratified into two broad categories. Type I tumors are predominantly driven by unopposed estrogen exposure, are typically low-grade, more frequently encountered, and generally associated with a favorable prognosis. In contrast, Type II tumors are largely estrogen-independent, characterized by high-grade histology, less frequent occurrence, and a comparatively poor clinical outcome (236). In women with endometrial cancer, alterations in the vaginal microbiota have been observed, characterized by the presence of specific bacterial taxa, including Firmicutes, Spirochaetes, Actinobacteria (e.g., Atopobium), and Proteobacteria (e.g., Bacteroides and Porphyromonas), often accompanied by an elevated vaginal pH (237). Notably, Atopobium and Porphyromonas have been shown to stimulate the release of proinflammatory cytokines, including IL-1α, IL-1β, IL-17α, and TNF-α (238).
Ovarian cancer (OC) represents the second most prevalent malignancy of the female reproductive system, following endometrial cancer, and predominantly arises in postmenopausal women. The disease primarily affects individuals aged 55-70 years, with incidence peaking between ages 55 and 59. Alarmingly, approximately 70% of ovarian cancer cases are diagnosed at advanced stages (FIGO stages III-IV), reflecting the insidious onset and lack of specific early clinical manifestations (239). Emerging evidence indicates a potential link between the gut microbiota and ovarian cancer. The gut microbial community has been shown to influence systemic inflammatory processes and modulate host immune responses, thereby shaping the ovarian tumor microenvironment and potentially contributing to disease initiation and progression (240). Microbiome diversity and richness within OC niches are markedly reduced, with certain taxa exhibiting relative enrichment compared to non-cancerous tissues (241-243). Notably, Propionibacterium acnes, Acetobacter, members of the phyla Firmicutes, Proteobacteria, and Fusobacterium demonstrate increased abundance, whereas Lactococcus is significantly diminished (159, 241-245). Several of these bacteria have been implicated in shaping a pro-tumorigenic inflammatory microenvironment by activating inflammatory signaling cascades and oxidative stress responses. By isolating and culturing specific strains, Huang et al. confirmed the overrepresentation of these genera and identified P. acnes as the predominant strain in OC. Functional assays further demonstrated its tumor-promoting role in epithelial ovarian cancer (EOC), wherein P. acnes activates the Hedgehog pathway and elevates proinflammatory mediators, including TNF-α and IL-1β (246). Additionally, iron-induced oxidative stress mediated by Acetobacter and Lactobacillus in clear-cell OC drives persistent inflammation, DNA damage, and oncogene activation, ultimately fostering tumor progression (247). However, whether these microbial effects are sufficient to initiate tumorigenesis or merely accelerate preexisting oncogenic processes remains a matter of debate. Notably, conflicting data exist regarding whether microbial-derived metabolites act as tumor suppressors or promoters, highlighting the context-dependent nature of host-microbiome interactions. The relationship between cancer and microorganisms, as described above, is illustrated in the figure below (Figure 3).
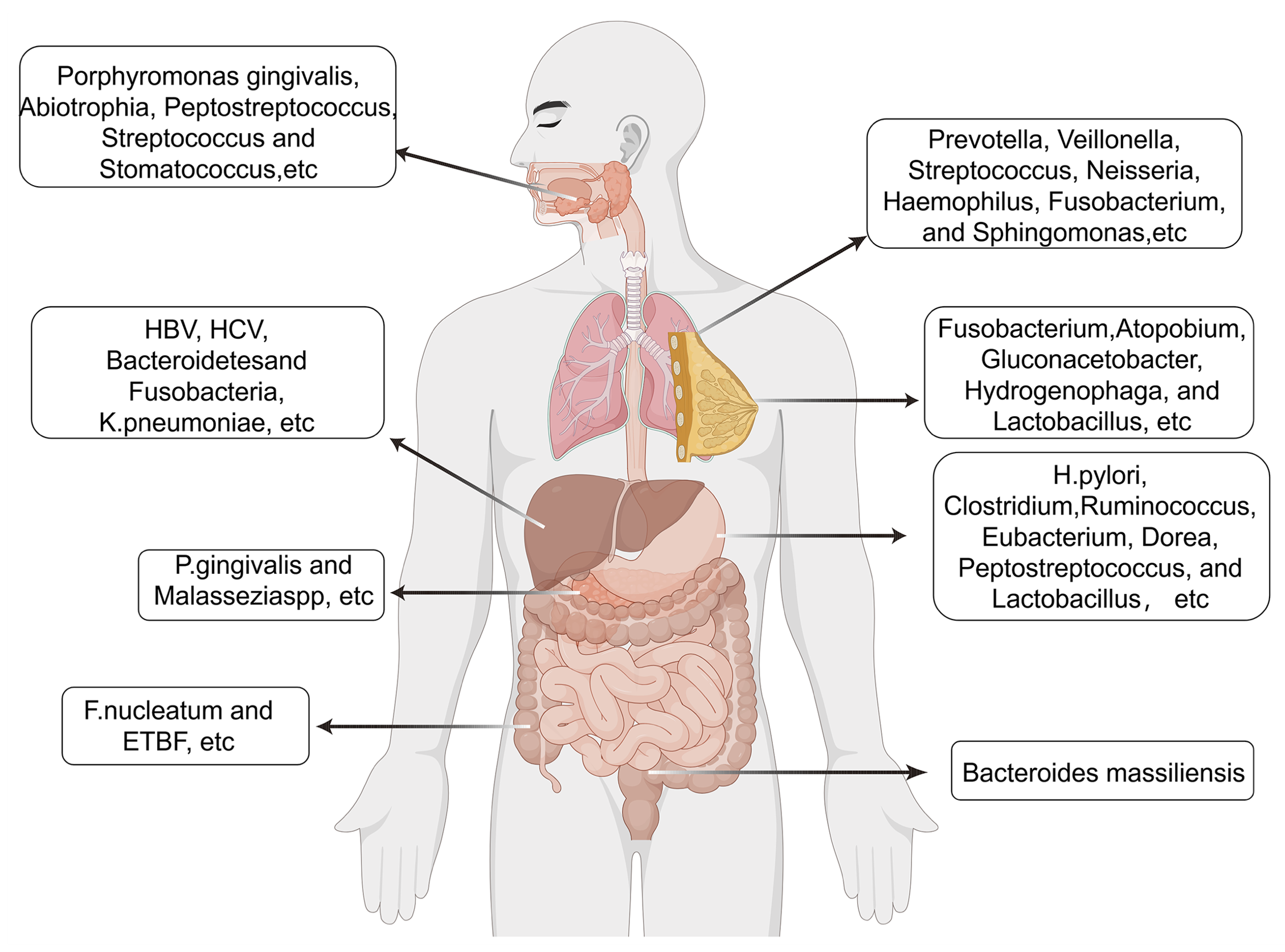
Figure 3. Microbiome signatures and clinical implications across major cancer types. The figure integrates distinct microbial taxa or communities associated with individual malignancies. Colored nodes denote microbial taxa enriched in specific tumors, while connecting lines represent shared or overlapping microbial associations. Icons indicate key biological effects, including inflammation, metabolic alteration, immune modulation, or direct oncogenic activity. This integrative framework highlights the potential of microbial profiles as diagnostic, prognostic, and therapeutic biomarkers across diverse cancer types.
3. Microbiome and Cancer Therapy
Probiotics are defined as live microorganisms that, when administered in sufficient amounts, confer health benefits to the host (248). They are widely used as standardized dietary supplements and are generally recognized as safe (249). A major mechanism through which probiotics exert beneficial effects is the production of short-chain fatty acids (SCFAs), particularly butyrate, generated by the fermentation of polysaccharides by species such as Clostridium butyricum and Akkermansia muciniphila (250). Butyrate has pleiotropic roles, including regulation of immune responses, modulation of intestinal hormone secretion, and regulation of lipid metabolism. For example, butyrate has been shown to induce apoptosis in colon cancer cell lines, suppressing tumor cell growth by upregulating the cyclin-dependent kinase inhibitor p57, thereby contributing to cell cycle arrest and tumor suppression (251). Prebiotics are defined as selectively fermented, non-digestible dietary fibers that promote the growth and activity of probiotic microorganisms. By maintaining intestinal microbial homeostasis and mitigating gut dysbiosis, prebiotics play a significant role in promoting host health. Their primary site of action is the colon, where they modulate resident populations of Lactobacilli and Bifidobacteria, thereby enhancing SCFA production. These SCFAs exert diverse physiological effects, including reinforcement of the gut epithelial and mucus barriers, regulation of immune responses, modulation of glucose and lipid metabolism, and influence on energy expenditure and satiety (252).
Beyond their local effects on epithelial integrity, SCFAs, notably acetate, propionate, and butyrate, play essential roles in systemic immune regulation and metabolic reprogramming. Butyrate acts as a histone deacetylase (HDAC) inhibitor, promoting the differentiation of regulatory T cells (Tregs) through enhanced FOXP3 expression and suppressing pro-inflammatory Th17 responses (253). SCFAs also regulate macrophage polarization, shifting M1-like inflammatory phenotypes toward M2-like, anti-inflammatory states via G-protein-coupled receptors (GPR41, GPR43) and downstream AMP-activated protein kinase (AMPK) signaling. Furthermore, by serving as energy substrates in colonocytes and tumor-associated immune cells, SCFAs influence metabolic rewiring, including modulation of glycolysis and fatty acid oxidation. Bile acids (BAs), another major class of microbiota-derived metabolites, exert equally profound effects on tumor biology. Primary BAs synthesized in the liver are converted into secondary BAs such as deoxycholic acid (DCA) and lithocholic acid (LCA) by intestinal bacteria (253). Collectively, SCFAs and BAs exemplify how microbial metabolites link gut microbial ecology with host immune-metabolic networks, influencing both tumor initiation and therapeutic response.
The immune system plays a central role in tumor surveillance and suppression, and strategies that harness its activity have become pivotal in cancer therapy. Among these, immunotherapy has emerged as a transformative treatment modality across diverse malignancies. In particular, immune checkpoint blockade (ICB) has gained prominence, employing monoclonal antibodies that target inhibitory pathways, such as programmed cell death protein 1 (PD-1), its ligand PD-L1, and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4). By releasing the brakes on T-cell activation, these agents enhance antitumor immune responses and have demonstrated durable clinical benefits in subsets of patients (254). Hua D et al. demonstrated that anti-PD-L1 therapy, when combined with Clostridium butyricum (CB) and Akkermansia muciniphila (AKK), markedly suppressed colitis-associated colorectal cancer (CRC) progression. This combination not only attenuated excessive activation of CD8⁺ T cells and macrophages within the inflammatory milieu but also enhanced CRC cell responsiveness to anti-PD-L1 treatment. Collectively, these findings suggest that CB and AKK exert direct antitumor effects, thereby improving the efficacy of immune checkpoint blockade and providing a promising therapeutic approach (255). Specific commensals, such as Bifidobacterium and Akkermansia muciniphila, enhance anti-PD-(L)1 or anti-CTLA-4 responses by activating dendritic cells, improving antigen presentation, and promoting the infiltration of cytotoxic CD8⁺ T cells into tumors. In contrast, broad-spectrum antibiotics or germ-free conditions markedly reduce ICI efficacy and alter the tumor immune microenvironment toward an immunosuppressive phenotype (Table 5).
Table 5. Representative studies on microbiome-ICI interactions.
| Study type | Cancer type |
Intervention/ Exposure |
Microbiome features | Immune effects | Clinical outcome |
|---|---|---|---|---|---|
| Preclinical | Melanoma (mouse) |
Bifidobacterium + anti-PD-L1 |
↑ DC activation, ↑ CD8⁺ T-cell infiltration |
↑ IFN-γ response | Enhanced tumor control |
| Preclinical | Multiple (mouse) | Broad-spectrum antibiotics before ICI | ↓ Microbial diversity | ↓ Antigen presentation, ↑ MDSCs | Reduced ICI efficacy |
| Observational | Melanoma/Lung/RCC | Baseline gut microbiome | ↑ Akkermansia, Faecalibacterium | ↑ Th1/CTL signatures | Improved ORR/PFS/OS |
| Interventional | Melanoma (phase I) |
FMT from responders + anti-PD-1 |
Microbiome shifted to responder-like pattern | ↑ T-cell activation | Partial responses in resistant |
| Interventional | RCC (early-phase) | SCFA-producing probiotic + ICI | ↑ Clostridium spp. abundance | Enhanced T-cell function | Preliminary PFS benefit (ongoing) |
Across melanoma, NSCLC, and renal cell carcinoma cohorts, higher baseline gut microbial diversity has been consistently associated with improved ICI responses and longer progression-free or overall survival. Enrichment of Akkermansia, Bifidobacterium, and Faecalibacterium species correlates with a greater Th1 and cytotoxic T-cell signature. In contrast, the dominance of oral-derived or potentially pathogenic taxa (e.g., Enterococcus faecalis) is linked to resistance. Concomitant exposure to antibiotics within ±60 days of ICI initiation reduces clinical benefit, while prolonged use of proton-pump inhibitors has been associated with microbiome perturbation and diminished outcomes (254). These associations underscore the microbiome as a potential predictive biomarker for immunotherapy efficacy.
Chemotherapy remains a cornerstone of cancer treatment; however, its bidirectional interactions with the host microbiome are increasingly recognized. On one hand, chemotherapeutic agents disrupt the intestinal microbial community, often exacerbating gastrointestinal toxicity and systemic complications, particularly in immunocompromised patients. Conversely, the microbiota can metabolize drugs, thereby modulating their pharmacokinetics, efficacy, and toxicity (161, 256-260). Emerging evidence highlights three principal roles of the gut microbiota in this context: enhancing therapeutic efficacy, augmenting antitumor activity, and mitigating adverse effects. In some cases, the microbiota itself may serve as a therapeutic target to mitigate chemotherapy-associated gastrointestinal toxicity (261). Fecal microbiota transplantation (FMT) is a therapeutic strategy in which functional microbiota derived from donor feces are introduced into the gastrointestinal (GI) tract of patients to alter and restore gut microbial composition. Initially, FMT was recognized for its remarkable efficacy in treating Clostridioides difficile infection (CDI) (262).
Since then, its clinical application has expanded considerably, offering new therapeutic avenues for a broad spectrum of diseases associated with dysbiosis of the gut microbiome (263). Although the precise molecular and cellular mechanisms underlying FMT remain incompletely elucidated, it is thought to involve direct interactions between the donor microbiota and the host, influencing mucosal barrier integrity, immune regulation, and systemic physiology. Preclinical studies further suggest that FMT accelerates recovery in chemotherapy-treated mice, underscoring its potential to restore microbial homeostasis, enhance chemotherapeutic efficacy, and mitigate inflammation and toxicity (264, 265) (Figure 4).
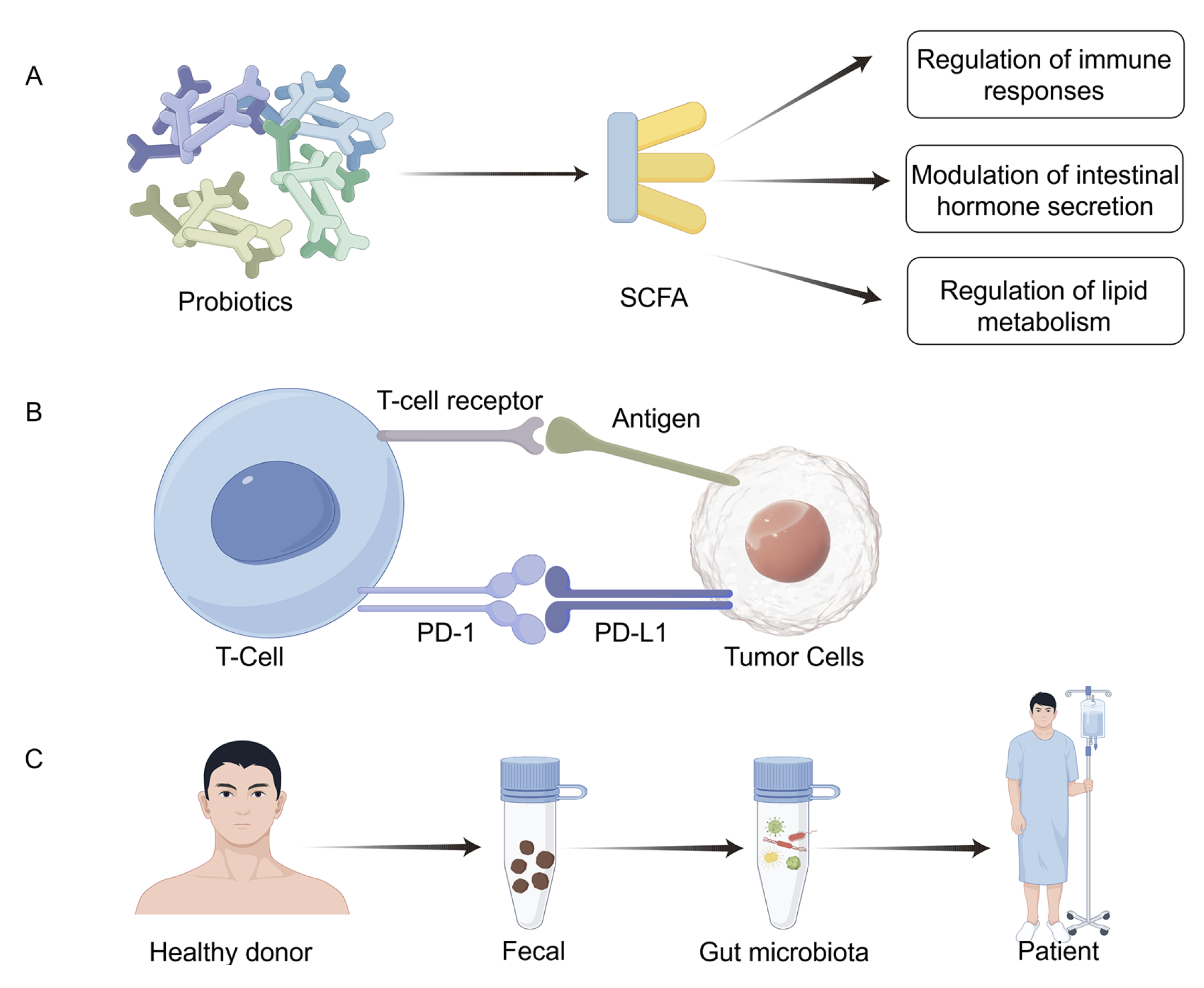
Figure 4. Microbiome modulation strategies in cancer prevention and therapy. The figure summarizes current and emerging strategies for modulating the microbiome in oncology. Approaches include antibiotics or bacteriophage therapy to deplete harmful taxa, probiotics and prebiotics to restore microbial balance, dietary interventions to optimize metabolite production, and fecal microbiota transplantation (FMT) to reconstitute a healthy microbial community. The lower panel illustrates how microbiome-targeted modulation can enhance responses to immunotherapy, chemotherapy, and radiotherapy. These interventions represent a translational bridge between microbial ecology and precision oncology (Table 6).
Table 6: Microbiome in Cancer Therapy
| Therapy | Microbial Influence | Key Findings | Clinical Implication |
|---|---|---|---|
|
Probiotics prebiotics |
Clostridium, Butyricum, Lactobacilli, Bifidobacteria (251, 252) | Butyrate's regulatory effects on immune response, intestinal hormone secretion, and lipid metabolism. Promotes resistance. | Improving gut health, boosting the immune system. |
| Immunotherapy | Clostridium butyricum and Akkermansia muciniphila (255) | Butyrate's regulatory effects on immune response, intestinal hormone secretion, and lipid metabolism. Promotes resistance. | Enhance antitumor immune responses. |
| FMT | Clostridioides difficile (266) | Restore microbial homeostasis, enhance chemotherapeutic efficacy, and attenuate inflammation and toxicity. |
Despite promising mechanistic and clinical findings, significant barriers hinder the translation of microbiome research into oncology practice. One major challenge is inter-individual variability; the human microbiome is highly dynamic and influenced by host genetics, diet, age, medication use, and comorbidities, all of which confound reproducibility across studies. Geographic and cultural differences further shape microbial composition, resulting in region-specific taxa that limit the generalizability of predictive biomarkers and therapeutic interventions identified in single populations. Additionally, the absence of standardized methodologies, including differences in sample collection (stool vs. tissue vs. urine), storage, sequencing depth, bioinformatic pipelines, and statistical normalization, remains a critical obstacle to meta-analysis and regulatory acceptance. Variability in analytical parameters can generate artificial discrepancies even when studying the same cancer type.
4. Clinical Implications of Microbiome Research in Oncology
The integration of microbiome science into clinical oncology presents transformative opportunities for diagnosis, prognostication, and personalized treatment. Microbial signatures from stool, tissue, or blood can serve as non-invasive biomarkers for early detection, risk stratification, and therapeutic monitoring. In parallel, understanding how specific taxa and metabolites influence immune activation, drug metabolism, and systemic inflammation enables the development of microbiome-informed precision medicine. For example, baseline gut diversity and enrichment of beneficial commensals correlate with improved outcomes in patients receiving immune checkpoint inhibitors, while dysbiosis can predict resistance or treatment-related toxicity. Consequently, microbiome modulation strategies, including dietary interventions, probiotics, prebiotics, FMT, and LBPs, are emerging as adjunctive tools to optimize therapeutic efficacy and minimize adverse effects. To translate these advances safely and effectively, prospective clinical trials with standardized sampling, longitudinal follow-up, and multi-omics integration are essential. Ultimately, incorporating microbiome assessment into routine oncology practice could refine patient selection, guide the development of combination therapies, and improve overall clinical outcomes.
Although mounting evidence connects microbial dysbiosis to carcinogenesis, much of the current literature remains correlative and heterogeneous. Variability in study design, sequencing methodology, and statistical control contributes to inconsistent results. Moreover, many mechanistic conclusions are drawn from single-species or animal models, which may not fully capture the complexity of human tumor-microbe ecosystems. Future research should integrate multi-omics, spatial, and temporal data to discern causal mechanisms and clarify whether the microbiome acts as a driver, a modulator, or merely a bystander in tumor evolution.
Discussion
Although substantial evidence links the microbiome to cancer development, most existing studies are associative rather than causal. Establishing causality between microbial alterations and oncogenesis remains a critical research priority. Future investigations should utilize longitudinal cohort designs, mechanistic experiments in gnotobiotic or organoid models, and rigorously controlled clinical interventions, such as fecal microbiota transplantation (FMT) or targeted microbial modulation, to clarify cause-and-effect relationships. Additionally, standardized sampling, contamination control, and multi-omics integration are essential for improving reproducibility and comparability across cohorts.
A significant challenge remains in translating microbiome research into clinical oncology. Although microbial signatures show promise as biomarkers for early detection and prediction of treatment response, their integration into precision oncology requires validated analytical pipelines, regulatory standards, and multidisciplinary collaboration. Bridging the gap between correlation and causation will ultimately allow the microbiome to transition from a descriptive hallmark of cancer to a targetable component of tumor biology and therapeutic innovation.
Several key research questions require systematic investigation: (1) Causation versus correlation: Most current findings are associative. Disentangling causal mechanisms necessitates longitudinal follow-up, interventional trials, and mechanistic validation in gnotobiotic or organoid models. Advanced causal inference methods, such as Mendelian randomization, may also clarify directionality. (2) Common versus cancer-specific microbial signatures: It remains uncertain whether shared microbial patterns drive carcinogenesis across multiple tumor types or if each malignancy possesses a unique microbiome configuration. Large-scale, cross-cancer microbiome atlases and meta-analyses are needed to address this issue. (3) Standardization and reproducibility: Methodological variability, including sampling strategies, sequencing depth, contamination control, and bioinformatics pipelines, continues to impede cross-study comparability. Establishing global standards for microbiome detection and reporting is essential to ensure data reliability. (4) Clinical translation: The application of microbiome insights to diagnostic and therapeutic practice will require validated biomarkers, safe and effective intervention strategies, and integrative models that combine microbial, genomic, and immunologic features. Rigorous clinical trials are necessary to determine how microbiome modulation can enhance responses to immunotherapy, chemotherapy, and targeted therapy. Addressing these questions will advance microbiome-oncology research from correlation-driven observation to a mechanistically informed and clinically actionable discipline.
References
1. DiMaio D, Emu B, Goodman AL, Mothes W, Justice A. Cancer microbiology. J Natl Cancer Inst. 2022;114(5):651-663. https://doi.org/10.1093/jnci/djab212
2. Wang Y, Zhang R, Pu Y, Wang D, Wang Y, Wu X, et al. Sample collection, DNA extraction, and library construction protocols of the human microbiome studies in the International Human Phenome Project. Phenomics. 2023;3(3):300-308. https://doi.org/10.1007/s43657-023-00097-y
3. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5(10):3111-3122. PMID: 26609491. PMCID: PMC4656734
4. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer compared with benign mass-like lesions. Lung Cancer. 2016;102:89-95. https://doi.org/10.1016/j.lungcan.2016.10.016
5. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198(9):1188-1198. https://doi.org/10.1164/rccm.201710-2118OC
6. Chu S, Cheng Z, Yin Z, Xu J, Wu F, Jin Y, et al. Airway Fusobacterium is associated with poor response to immunotherapy in lung cancer. Onco Targets Ther. 2022;15:201-213. https://doi.org/10.2147/OTT.S348382
7. Derosa L, Routy B, Thomas AM, Iebba V, Zalcman G, Friard S, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med. 2022;28(2):315-324. https://doi.org/10.1038/s41591-021-01655-5
8. Teng L, Wang K, Chen W, Wang YS, Bi L. HYR-2 plays an anti-lung cancer role by regulating PD-L1 and Akkermansia muciniphila. Pharmacol Res. 2020;160:105086. https://doi.org/10.1016/j.phrs.2020.105086
9. Zhang Z, Liu D, Liu S, Zhang S, Pan Y. The role of Porphyromonas gingivalis outer membrane vesicles in periodontal disease and related systemic diseases. Front Cell Infect Microbiol. 2020;10:585917. https://doi.org/10.3389/fcimb.2020.585917
10. Pignatelli P, Nuccio F, Piattelli A, Curia MC. The role of Fusobacterium nucleatum in oral and colorectal carcinogenesis. Microorganisms. 2023;11(9):2358. https://doi.org/10.3390/microorganisms11092358
11. Bakhti SZ, Latifi-Navid S. Interplay and cooperation of Helicobacter pylori and gut microbiota in gastric carcinogenesis. BMC Microbiol. 2021;21(1):258. https://doi.org/10.1186/s12866-021-02315-x
12. Alipour M. Molecular mechanism of Helicobacter pylori-induced gastric cancer. J Gastrointest Cancer. 2021;52(1):23-30. https://doi.org/10.1007/s12029-020-00518-5
13. Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, et al. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. 2021;11(10):4945-4956. https://doi.org/10.7150/thno.55209
14. Rocken C. Predictive biomarkers in gastric cancer. J Cancer Res Clin Oncol. 2023;149(1):467-481. https://doi.org/10.1007/s00432-022-04408-0
15. Fletcher AA, Kelly MS, Eckhoff AM, Allen PJ. Revisiting the intrinsic mycobiome in pancreatic cancer. Nature. 2023;620(7972):E1-E6. https://doi.org/10.1038/s41586-023-06292-1
16. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. 2022;14(1):2073785. https://doi.org/10.1080/19490976.2022.2073785
17. Hayashi M, Ikenaga N, Nakata K, Luo H, Zhong P, Date S, et al. Intratumor Fusobacterium nucleatum promotes the progression of pancreatic cancer via the CXCL1–CXCR2 axis. Cancer Sci. 2023;114(9):3666-3678. https://doi.org/10.1111/cas.15901
18. Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023;31(2):159-172. https://doi.org/10.1016/j.tim.2022.08.010
19. Jasemi S, Molicotti P, Fais M, Cossu I, Simula ER, Sechi LA. Biological mechanisms of enterotoxigenic Bacteroides fragilis toxin: linking inflammation, colorectal cancer, and clinical implications. Toxins (Basel). 2025;17(6):305. https://doi.org/10.3390/toxins17060305
20. Chen B, Ramazzotti D, Heide T, Spiteri I, Fernandez-Mateos J, James C, et al. Contribution of pks(+) E. coli mutations to colorectal carcinogenesis. Nat Commun. 2023;14(1):7827. https://doi.org/10.1038/s41467-023-43329-5
21. Huang C, Mei S, Zhang X, Tian X. Inflammatory milieu related to dysbiotic gut microbiota promotes tumorigenesis of hepatocellular carcinoma. J Clin Gastroenterol. 2023;57(8):782-788. https://doi.org/10.1097/MCG.0000000000001883
22. Shi Q, Wang J, Zhou M, Zheng R, Zhang X, Liu B. Gut Lactobacillus contributes to the progression of breast cancer by affecting the anti-tumor activities of immune cells in the TME of tumor-bearing mice. Int Immunopharmacol. 2023;124(Pt B):111039. https://doi.org/10.1016/j.intimp.2023.111039
23. Peng F, Hu M, Su Z, Hu L, Guo L, Yang K. Intratumoral microbiota as a target for advanced cancer therapeutics. Adv Mater. 2024;36(38):2405331. https://doi.org/10.1002/adma.202405331
24. Barrett M, Hand CK, Shanahan F, Murphy T, O’Toole PW. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer. 2020;6(4):277-287. https://doi.org/10.1016/j.trecan.2020.01.019
25. Jiang M, Yang Z, Dai J, Wu T, Jiao Z, Yu Y, et al. Intratumor microbiome: selective colonization in the tumor microenvironment and a vital regulator of tumor biology. MedComm (2020). 2023;4(5):e376. https://doi.org/10.1002/mco2.376
26. Joo JE, Chu YL, Georgeson P, Walker R, Mahmood K, Clendenning M, et al. Intratumoral presence of the genotoxic gut bacteria pks(+) E. coli, enterotoxigenic Bacteroides fragilis, and Fusobacterium nucleatum and their association with clinicopathological and molecular features of colorectal cancer. Br J Cancer. 2024;130(5):728-740. https://doi.org/10.1038/s41416-023-02554-x
27. Wong CC, Yu J. pks(+) E. coli adhesins—the fine line between good and evil. Cell Host Microbe. 2025;33(1):1-3. https://doi.org/10.1016/j.chom.2024.12.007
28. Cao Y, Oh J, Xue M, Huh WJ, Wang J, González-Hernández JA, et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science. 2022;378(6618):eabm3233. https://doi.org/10.1126/science.abm3233
29. Khan S. Potential role of Escherichia coli DNA mismatch-repair proteins in colon cancer. Crit Rev Oncol Hematol. 2015;96(3):475-482. https://doi.org/10.1016/j.critrevonc.2015.05.002
30. Santos JC, Brianti MT, Almeida VR, Ortega MM, Fischer W, Haas R, et al. Helicobacter pylori infection modulates the expression of miRNAs associated with the DNA mismatch repair pathway. Mol Carcinog. 2017;56(4):1372-1379. https://doi.org/10.1002/mc.22590
31. Khan FH, Dervan E, Bhattacharyya DD, McAuliffe JD, Miranda KM, Glynn SA. The role of nitric oxide in cancer: master regulator or NOt? Int J Mol Sci. 2020;21(24):9393. https://doi.org/10.3390/ijms21249393
32. Zhao Y, Ye X, Xiong Z, Ihsan A, Ares I, Martínez M, et al. Cancer metabolism: the role of ROS in DNA damage and induction of apoptosis in cancer cells. Metabolites. 2023;13(7):796. https://doi.org/10.3390/metabo13070796
33. DeCaprio JA. Molecular pathogenesis of Merkel cell carcinoma. Annu Rev Pathol. 2021;16:69-91. https://doi.org/10.1146/annurev-pathmechdis-012419-032817
34. Torre-Castro J, Rodríguez M, Alonso-Alonso R, Mendoza Cembranos MD, Díaz-Alejo JF, Rebollo-González M, et al. LT and SOX9 expression are associated with gene sets that distinguish Merkel cell polyomavirus (MCPyV)-positive and MCPyV-negative Merkel cell carcinoma. Br J Dermatol. 2024;190(6):876-884. https://doi.org/10.1093/bjd/ljae033
35. Fernandes Q, Inchakalody VP, Bedhiafi T, Mestiri S, Taib N, Uddin S, et al. Chronic inflammation and cancer; the two sides of a coin. Life Sci. 2024;338:122390. https://doi.org/10.1016/j.lfs.2023.122390
36. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:1533033819867354. https://doi.org/10.1177/1533033819867354
37. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254-258. https://doi.org/10.1038/nature11465
38. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7-34. https://doi.org/10.3322/caac.21551
39. Tang Z, Liang D, Deubler EL, Sarnat JA, Chow SS, Diver WR, et al. Lung cancer metabolomics: a pooled analysis in the Cancer Prevention Studies. BMC Med. 2024;22(1):262. https://doi.org/10.1186/s12916-024-03473-1
40. Rodríguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Treat Res. 2016;170:25-46. https://doi.org/10.1007/978-3-319-40389-2_2
41. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018;24(13):3528-3538. https://doi.org/10.1016/j.celrep.2018.08.090
42. Kaderbhai C, Richard C, Fumet JD, Aarnink A, Foucher P, Coudert B, et al. Antibiotic use does not appear to influence response to nivolumab. Anticancer Res. 2017;37(6):3195-3200. https://doi.org/10.21873/anticanres.11680
43. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957-963. https://doi.org/10.1164/rccm.201104-0655OC
44. Pilette C, Ouadrhiri Y, Godding V, Vaerman JP, Sibille Y. Lung mucosal immunity: immunoglobulin-A revisited. Eur Respir J. 2001;18(3):571-588. https://doi.org/10.1183/09031936.01.00228801
45. Toma I, Siegel MO, Keiser J, Yakovleva A, Kim A, Davenport L, et al. Single-molecule long-read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol. 2014;52(11):3913-3921. https://doi.org/10.1128/JCM.01678-14
46. McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, et al. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365(17):1567-1575. https://doi.org/10.1056/NEJMoa1106955
47. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2):e16384. https://doi.org/10.1371/journal.pone.0016384
48. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. https://doi.org/10.1371/journal.pone.0008578
49. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep. 2019;9(1):12838. https://doi.org/10.1038/s41598-019-49195-w
50. Chung KF. Airway microbial dysbiosis in asthmatic patients: a target for prevention and treatment? J Allergy Clin Immunol. 2017;139(4):1071-1081. https://doi.org/10.1016/j.jaci.2017.02.004
51. Hewitt RJ, Lloyd CM. Regulation of immune responses by the airway epithelial cell landscape. Nat Rev Immunol. 2021;21(6):347-362. https://doi.org/10.1038/s41577-020-00477-9
52. Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, et al. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int J Biol Sci. 2019;15(11):2381-2392. https://doi.org/10.7150/ijbs.35980
53. Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. 2019;9:112. https://doi.org/10.3389/fcimb.2019.00112
54. Zhang WQ, Zhao SK, Luo JW, Dong XP, Hao YT, Li H, et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res. 2018;10(10):3171-3185. https://pmc.ncbi.nlm.nih.gov/articles/PMC6220220/
55. Wang H, Hu J, Ma Y, Abulimiti Y, Zhou Y. Lung commensal bacteria promote lung cancer progression through NK cell-mediated immunosuppressive microenvironment. Int J Med Sci. 2025;22(5):1039-1051. https://doi.org/10.7150/ijms.107026
56. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R, et al. Specific gut microbiome signature predicts early-stage lung cancer. Gut Microbes. 2020;11(4):1030-1042. https://doi.org/10.1080/19490976.2020.1737487
57. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6(2):471-474. https://doi.org/10.1038/ismej.2011.104
58. Qiao D, Wang Z, Lu Y, Wen X, Li H, Zhao H. A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am J Cancer Res. 2014;5(1):423-432. https://pmc.ncbi.nlm.nih.gov/articles/PMC4300720/
59. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17(1):163. https://doi.org/10.1186/s13059-016-1021-1
60. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957-963. https://doi.org/10.1164/rccm.201104-0655OC
61. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. https://doi.org/10.1371/journal.pone.0008578
62. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821-830. https://doi.org/10.1513/AnnalsATS.201501-029OC
63. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6(2):471-474. https://doi.org/10.1038/ismej.2011.104
64. Cameron SJ, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One. 2017;12(5):e0177062. https://doi.org/10.1371/journal.pone.0177062
65. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821-830. https://doi.org/10.1513/AnnalsATS.201501-029OC
66. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. https://doi.org/10.1038/nmicrobiol.2016.31
67. Oriano M, Gramegna A, Terranova L, Sotgiu G, Sulaiman I, Ruggiero L, et al. Sputum neutrophil elastase associates with microbiota and Pseudomonas aeruginosa in bronchiectasis. Eur Respir J. 2020;56(4):2000769. https://doi.org/10.1183/13993003.00769-2020
68. Tsay JC, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198(9):1188-1198. https://doi.org/10.1164/rccm.201710-2118OC
69. Mac Aogáin M, Dicker AJ, Mertsch P, Chotirmall SH. Infection and the microbiome in bronchiectasis. Eur Respir Rev. 2024;33(173):240038. https://doi.org/10.1183/16000617.0038-2024
70. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998-1013.e16. https://doi.org/10.1016/j.cell.2018.12.040
71. Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488-500. https://doi.org/10.1007/s13238-018-0548-1
72. Yamashita Y, Takeshita T. The oral microbiome and human health. J Oral Sci. 2017;59(2):201-206. https://doi.org/10.2334/josnusd.16-0856
73. Santacroce L, Passarelli PC, Azzolino D, Bottalico L, Charitos IA, Cazzolla AP, Colella M, Topi S, Garcia-Godoy F, D’Addona A. Oral microbiota in human health and disease: a perspective. Exp Biol Med (Maywood). 2023;248(15):1288-1301. https://doi.org/10.1177/15353702231187645
74. Kilian M. The oral microbiome—friend or foe? Eur J Oral Sci. 2018;126(Suppl 1):5-12. https://doi.org/10.1111/eos.12527
75. Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777-6787. https://doi.org/10.1158/0008-5472.CAN-17-1296
76. Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95(7):569-574. https://doi.org/10.1111/j.1349-7006.2004.tb02488.x
77. Snider EJ, Compres G, Freedberg DE, Giddins MJ, Khiabanian H, Lightdale CJ, et al. Barrett’s esophagus is associated with a distinct oral microbiome. Clin Transl Gastroenterol. 2018;9(3):e135. https://doi.org/10.1038/s41424-018-0005-8
78. Vogtmann E, Han Y, Caporaso JG, Bokulich N, Mohamadkhani A, Moayyedkazemi A, et al. Oral microbial community composition is associated with pancreatic cancer: a case-control study in Iran. Cancer Med. 2020;9(2):797-806. https://doi.org/10.1002/cam4.2660
79. Del Castillo E, Meier R, Chung M, Koestler DC, Chen T, Paster BJ, et al. The microbiomes of pancreatic and duodenum tissue overlap and are highly subject-specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol Biomarkers Prev. 2019;28(2):370-383. https://doi.org/10.1158/1055-9965.EPI-18-0542
80. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454-1463. https://doi.org/10.1136/gutjnl-2017-314814
81. Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122-128. https://doi.org/10.4103/jomfp.JOMFP_304_18
82. Demmitt BA, Corley RP, Huibregtse BM, Keller MC, Hewitt JK, McQueen MB, et al. Genetic influences on the human oral microbiome. BMC Genomics. 2017;18(1):659. https://doi.org/10.1186/s12864-017-4008-8
83. Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162(2):22-38. https://doi.org/10.1016/j.imlet.2014.08.017
84. Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 2018;3(6):e00187-18. https://doi.org/10.1128/mSystems.00187-18
85. Oren A, Garrity GM. Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. 2021;71(10):005056. https://doi.org/10.1099/ijsem.0.005056
86. McLean JS. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:98. https://doi.org/10.3389/fcimb.2014.00098
87. Baker JL, Mark Welch JL, Kauffman KM, McLean JS, He X. The oral microbiome: diversity, biogeography and human health. Nat Rev Microbiol. 2024;22(2):89-104. https://doi.org/10.1038/s41579-023-00963-6
88. Diaz P, Dongari-Bagtzoglou A. Critically appraising the significance of the oral mycobiome. J Dent Res. 2021;100(2):133-140. https://doi.org/10.1177/0022034520956975
89. Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. https://doi.org/10.1186/1471-2164-14-623
90. Turner SA, Butler G. The Candida pathogenic species complex. Cold Spring Harb Perspect Med. 2014;4(9):a019778. https://doi.org/10.1101/cshperspect.a019778
91. Ahmad KM, Kokošar J, Guo X, Gu Z, Ishchuk OP, Piskur J. Genome structure and dynamics of the yeast pathogen Candida glabrata. FEMS Yeast Res. 2014;14(4):529-535. https://doi.org/10.1111/1567-1364.12145
92. Hong BY, Hoare A, Cardenas A, Dupuy A, Choquette L, Salner A, et al. The salivary mycobiome contains 2 ecologically distinct mycotypes. J Dent Res. 2020;99(6):730-738. https://doi.org/10.1177/0022034520915879
93. Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9(3):e90899. https://doi.org/10.1371/journal.pone.0090899
94. Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10(3):e1003933. https://doi.org/10.1371/journal.ppat.1003933
95. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. https://doi.org/10.3322/caac.21660
96. Irfan M, Delgado RZR, Frias-Lopez J. The oral microbiome and cancer. Front Immunol. 2020;11:591088. https://doi.org/10.3389/fimmu.2020.591088
97. Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2020;9:476. https://doi.org/10.3389/fcimb.2019.00476
98. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3(4):209-215. https://doi.org/10.4248/ijos11075
99. Sayehmiri F, Sayehmiri K, Asadollahi K, Soroush S, Bogdanovic L, Jalilian FA, et al. The prevalence rate of Porphyromonas gingivalis and its association with cancer: a systematic review and meta-analysis. Int J Immunopathol Pharmacol. 2015;28(2):160-167. https://doi.org/10.1177/0394632015586144
100. Wen L, Mu W, Lu H, Wang X, Fang J, Jia Y, et al. Porphyromonas gingivalis promotes oral squamous cell carcinoma progression in an immune microenvironment. J Dent Res. 2020;99(6):666-675. https://doi.org/10.1177/0022034520909312
101. Rai AK, Panda M, Das AK, Rahman T, Das R, Das K, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. 2021;203(1):137-152. https://doi.org/10.1007/s00203-020-02011-w
102. Mammen MJ, Scannapieco FA, Sethi S. Oral–lung microbiome interactions in lung diseases. Periodontol 2000. 2020;83(1):234-241. https://doi.org/10.1111/prd.12301
103. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59-64. https://doi.org/10.1038/nature13568
104. Acharya C, Sahingur SE, Bajaj JS. Microbiota, cirrhosis, and the emerging oral–gut–liver axis. JCI Insight. 2017;2(19):e94416. https://doi.org/10.1172/jci.insight.94416
105. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359-365. https://doi.org/10.1126/science.aan4526
106. Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182(2):447-462.e14. https://doi.org/10.1016/j.cell.2020.05.048
107. Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215-225. https://doi.org/10.1016/j.chom.2016.07.006
108. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case–control study. Gut. 2018;67(1):120-127. https://doi.org/10.1136/gutjnl-2016-312580
109. Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjonneland A, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62(12):1764-1770. https://doi.org/10.1136/gutjnl-2012-303006
110. Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. https://doi.org/10.7717/peerj.1373
111. Karpiński TM. The microbiota and pancreatic cancer. Gastroenterol Clin North Am. 2019;48(3):447-464. https://doi.org/10.1016/j.gtc.2019.04.008
112. Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209-7220. https://doi.org/10.18632/oncotarget.3109
113. Wei AL, Li M, Li GQ, Wang X, Hu WM, Li ZL, et al. Oral microbiome and pancreatic cancer. World J Gastroenterol. 2020;26(48):7679-7692. https://doi.org/10.3748/wjg.v26.i48.7679
114. Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582-588. https://doi.org/10.1136/gutjnl-2011-300784
115. Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5(12):1749-1768. https://doi.org/doi:10.1001/jamaoncol.2019.2996
116. Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23(4):713-739. https://doi.org/10.1128/CMR.00011-10
117. Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, et al. Clinical significance of four molecular subtypes of gastric cancer identified by The Cancer Genome Atlas Project. Clin Cancer Res. 2017;23(15):4441-4449. https://doi.org/10.1158/1078-0432.CCR-16-2211
118. Mommersteeg MC, Yu J, Peppelenbosch MP, Fuhler GM. Genetic host factors in Helicobacter pylori-induced carcinogenesis: emerging new paradigms. Biochim Biophys Acta Rev Cancer. 2018;1869(1):42-52. https://doi.org/10.1016/j.bbcan.2017.11.003
119. Castano-Rodríguez N, Kaakoush NO, Mitchell HM. Pattern-recognition receptors and gastric cancer. Front Immunol. 2014;5:336. https://doi.org/10.3389/fimmu.2014.00336
120. Moss SF. The clinical evidence linking Helicobacter pylori to gastric cancer. Cell Mol Gastroenterol Hepatol. 2017;3(2):183-191. https://doi.org/10.1016/j.jcmgh.2016.12.001
121. Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720-741. https://doi.org/10.1128/CMR.10.4.720
122. Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type gastric cancer. Sci Rep. 2014;4:4202. https://doi.org/10.1038/srep04202
123. Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407-416. https://doi.org/10.1111/hel.12145
124. Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7(1):15957. https://doi.org/10.1038/s41598-017-16289-2
125. Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7(3):e33418. https://doi.org/10.1371/journal.pone.0033418
126. Serban DE. Gastrointestinal cancers: influence of gut microbiota, probiotics and prebiotics. Cancer Lett. 2014;345(2):258-270. https://doi.org/10.1016/j.canlet.2013.08.013
127. Kidd M, Modlin IM. A century of Helicobacter pylori: paradigms lost–paradigms regained. Digestion. 1998;59(1):1-15. https://doi.org/10.1159/000007461
128. Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;321(8336):1273-1275. https://doi.org/10.1016/S0140-6736(83)92719-8
129. Llorca L, Pérez-Pérez G, Urruzuno P, Martinez MJ, Iizumi T, Gao Z, et al. Characterization of the gastric microbiota in a pediatric population according to Helicobacter pylori status. Pediatr Infect Dis J. 2017;36(2):173-178. https://doi.org/10.1097/INF.0000000000001383
130. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688-693. https://doi.org/10.1038/sj.embor.7400731
131. Minaga K, Watanabe T, Kamata K, Asano N, Kudo M. Nucleotide-binding oligomerization domain 1 and Helicobacter pylori infection: a review. World J Gastroenterol. 2018;24(16):1725-1733. https://doi.org/10.3748/wjg.v24.i16.1725
132. Yang J, Zhou X, Liu X, Ling Z, Ji F. Role of the gastric microbiome in gastric cancer: from carcinogenesis to treatment. Front Microbiol. 2021;12:641322. https://doi.org/10.3389/fmicb.2021.641322
133. Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, et al. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9(1):19. https://doi.org/10.1038/s41572-023-00431-8
134. Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol. 2017;112(2):212-239. https://doi.org/10.1038/ajg.2016.563
135. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859-904. https://doi.org/10.1152/physrev.00045.2009
136. Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer J. 2014;20(3):211-216. https://doi.org/10.1097/PPO.0000000000000043
137. Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863-1873. https://doi.org/10.1053/j.gastro.2009.01.073
138. Szabò I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, et al. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18(20):5517-5527. https://doi.org/10.1093/emboj/18.20.5517
139. Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, et al. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. J Pathol. 2007;213(2):140-151. https://doi.org/10.1002/path.2218
140. Bimela JS, Nanfack AJ, Yang P, Dai S, Kong XP, Torimiro JN, et al. Antiretroviral imprints and genomic plasticity of HIV-1 pol in non-clade B: implications for treatment. Front Microbiol. 2022;12:812391. https://doi.org/10.3389/fmicb.2021.812391
141. Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185(20):3807-3822.e12. https://doi.org/10.1016/j.cell.2022.09.015
142. Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X, et al. Integrative identification of Epstein–Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology. 2014;147(6):1350-1362.e4. https://doi.org/10.1053/j.gastro.2014.08.036
143. Yang J, Liu Z, Zeng B, Hu G, Gan R. Epstein–Barr virus-associated gastric cancer: a distinct subtype. Cancer Lett. 2020;495:191-199. https://doi.org/10.1016/j.canlet.2020.09.019
144. Sun K, Jia K, Lv H, Wang SQ, Wu Y, Lei H, et al. EBV-positive gastric cancer: current knowledge and future perspectives. Front Oncol. 2020;10:583463. https://doi.org/10.3389/fonc.2020.583463
145. Niedźwiedzka-Rystwej P, Grywalska E, Hrynkiewicz R, Wolacewicz M, Becht R, Roliński J. The double-edged sword role of viruses in gastric cancer. Cancers (Basel). 2020;12(6):1680. https://doi.org/10.3390/cancers12061680
146. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33. https://doi.org/10.3322/caac.21654
147. Ro C, Chai W, Yu VE, Yu R. Pancreatic neuroendocrine tumors: biology, diagnosis, and treatment. Chin J Cancer. 2013;32(6):312-324. https://doi.org/10.5732/cjc.012.10295
148. Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363(9414):1049-1057. https://doi.org/10.1016/S0140-6736(04)15841-8149. Cruz MS, Tintelnot J, Gagliani N. Roles of microbiota in pancreatic cancer development and treatment. Gut Microbes. 2024;16(1):2320280. https://doi.org/10.1080/19490976.2024.2320280
150. Kaźmierczak-Siedlecka K, Dvorák A, Folwarski M, Daca A, Przewlócka K, Makarewicz W. Fungal gut microbiota dysbiosis and its role in colorectal, oral, and pancreatic carcinogenesis. Cancers (Basel). 2020;12(5):1326. https://doi.org/10.3390/cancers12051326
151. Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18(1):97. https://doi.org/10.1186/s12943-019-1008-0
152. Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2015;12(2):77-87. https://doi.org/10.1038/nrgastro.2014.188
153. Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8(6):e66019. https://doi.org/10.1371/journal.pone.0066019
154. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264-267. https://doi.org/10.1038/s41586-019-1608-2
155. Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep. 2015;5:7980. https://doi.org/10.1038/srep07980
156. Li S, Fuhler GM, Bn N, Jose T, Bruno MJ, Peppelenbosch MP, et al. Pancreatic cyst fluid harbors a unique microbiome. Microbiome. 2017;5(1):147. https://doi.org/10.1186/s40168-017-0363-6
157. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403-416. https://doi.org/10.1158/2159-8290.CD-17-1134
158. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512-516. https://doi.org/10.1038/nature24462
159. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973-980. https://doi.org/10.1126/science.aay9189
160. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795-806.e12. https://doi.org/10.1016/j.cell.2019.07.008
161. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156-1160. https://doi.org/10.1126/science.aah5043
162. Fiorillo L, Cervino G, Laino L, D’Amico C, Mauceri R, Tozum TF, et al. Porphyromonas gingivalis, periodontal and systemic implications: a systematic review. Dent J (Basel). 2019;7(4):114. https://doi.org/10.3390/dj7040114
163. Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels, and orodigestive cancer mortality. Carcinogenesis. 2012;33(5):1055-1058. https://doi.org/10.1093/carcin/bgs112
164. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case–control study. Gut. 2018;67(1):120-127. https://doi.org/10.1136/gutjnl-2016-312580
165. Huang J, Roosaar A, Axéll T, Ye W. A prospective cohort study on poor oral hygiene and pancreatic cancer risk. Int J Cancer. 2016;138(2):340-347. https://doi.org/10.1002/ijc.29710
166. Jin Y, Gao H, Chen H, Wang J, Chen M, Li G, et al. Identification and impact of hepatitis B virus DNA and antigens in pancreatic cancer tissues and adjacent non-cancerous tissues. Cancer Lett. 2013;335(2):447-454. https://doi.org/10.1016/j.canlet.2013.03.001
167. Shimoda T, Shikata T, Karasawa T, Tsukagoshi S, Yoshimura M, Sakurai I. Light microscopic localization of hepatitis B virus antigens in the human pancreas: possibility of multiplication of hepatitis B virus in the human pancreas. Gastroenterology. 1981;81(6):998-1005. https://doi.org/10.1016/S0016-5085(81)80004-2
168. Arafa A, Eshak ES, Abdel Rahman TA, Anwar MM. Hepatitis C virus infection and risk of pancreatic cancer: a meta-analysis. Cancer Epidemiol. 2020;65:101691. https://doi.org/10.1016/j.canep.2020.101691
169. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7-33. https://doi.org/10.3322/caac.21708
170. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322-340. https://doi.org/10.1053/j.gastro.2019.06.048
171. Wang J, Chen W-D, Wang Y-D. The relationship between gut microbiota and inflammatory diseases: the role of macrophages. Front Microbiol. 2020;11:1065. https://doi.org/10.3389/fmicb.2020.01065
172. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223-237. https://doi.org/10.1038/s41575-019-0258-z
173. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299-306. https://doi.org/10.1101/gr.126516.111
174. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207-215. https://doi.org/10.1016/j.chom.2013.07.007
175. Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res (Phila). 2017;10(7):398-409. https://doi.org/10.1158/1940-6207.CAPR-16-0178
176. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195-206. https://doi.org/10.1016/j.chom.2013.07.012
177. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590. https://doi.org/10.1038/s41598-017-11237-6
178. Guinney J, Dienstmann R, Wang X, De Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350-1356. https://doi.org/10.1038/nm.3967
179. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443-1448. https://doi.org/10.1126/science.aal5240
180. Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22(2):349-369. https://doi.org/10.1128/CMR.00053-08
181. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, Shields CED, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018; 23(2):203-214.e5. https://doi.org/10.1016/j.chom.2018.01.007
182. Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016-1022. https://doi.org/10.1038/nm.2015
183. Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe JL, et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5(10):1098-1109. https://doi.org/10.1158/2159-8290.CD-15-0447
184. DeDecker L, Coppedge B, Avelar-Barragan J, Karnes W, Whiteson K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes. 2021;13(1):1-12. https://doi.org/10.1080/19490976.2020.1854641
185. Goodwin AC, Shields CED, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354-15359. https://doi.org/10.1073/pnas.1010203108
186. Allen J, Hao S, Sears CL, Timp W. Epigenetic changes induced by Bacteroides fragilis toxin. Infect Immun. 2019;87(6):e00447-18. https://doi.org/10.1128/IAI.00447-18
187. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289-300. https://doi.org/10.1136/gutjnl-2018-317200
188. Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848-851. https://doi.org/10.1126/science.1127059
189. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3(12):1683-1691. https://doi.org/10.1001/jamaoncol.2017.3055
190. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589-604. https://doi.org/10.1038/s41575-019-0186-y
191. Thilakarathna W, Rupasinghe HPV, Ridgway ND. Mechanisms by which probiotic bacteria attenuate the risk of hepatocellular carcinoma. Int J Mol Sci. 2021;22(5):2606. https://doi.org/10.3390/ijms22052606
192. Zhang N, Gou Y, Liang S, Chen N, Liu Y, He Q, et al. Dysbiosis of gut microbiota promotes hepatocellular carcinoma progression by regulating the immune response. J Immunol Res. 2021;2021:4973589. https://doi.org/10.1155/2021/4973589
193. Zheng R, Wang G, Pang Z, Ran N, Gu Y, Guan X, et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020;9(12):4232-4250. https://doi.org/10.1002/cam4.3045
194. Wang X, Fang Y, Liang W, Cai Y, Wong CC, Wang J, et al. Gut-liver translocation of pathogen Klebsiella pneumoniae promotes hepatocellular carcinoma in mice. Nat Microbiol. 2025;10(1):169-184. https://doi.org/10.1038/s41564-024-01890-9
195. Song Y, Lau HC, Zhang X, Yu J. Bile acids, gut microbiota, and therapeutic insights in hepatocellular carcinoma. Cancer Biol Med. 2023;21(2):144-162. https://doi.org/10.20892/j.issn.2095-3941.2023.0394
196. Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes. 2014;5(4):441-445. https://doi.org/10.4161/gmic.29599
197. Shen S, Khatiwada S, Behary J, Kim R, Zekry A. Modulation of the gut microbiome to improve clinical outcomes in hepatocellular carcinoma. Cancers (Basel). 2022;14(9):2099. https://doi.org/10.3390/cancers14092099
198. Ram AK, Pottakat B, Vairappan B. Increased systemic zonula occludens1 associated with inflammation and independent biomarker in patients with hepatocellular carcinoma. BMC Cancer. 2018;18(1):572. https://doi.org/10.1186/s12885-018-4484-5
199. Guo S, Al-Sadi R, Said HM, Ma TY. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am J Pathol. 2013;182(2):375-387. https://doi.org/10.1016/j.ajpath.2012.10.014
200. Behary J, Raposo AE, Amorim NML, Zheng H, Gong L, McGovern E, et al. Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2⁻/⁻ mouse model. BMC Microbiol. 2021;21(1):113. https://doi.org/10.1186/s12866-021-02171-9
201. Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25(1):79. https://doi.org/10.1186/s12929-018-0483-8
202. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504-516. https://doi.org/10.1016/j.ccr.2012.02.007
203. Yu LX, Yan HX, Liu Q, Yang W, Wu HP, Dong W, et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52(4):1322-1333. https://doi.org/10.1002/hep.23845
204. Ranaee M, Torabi H, Azhganzad N, Shirini K, Hosseini AS, Hajian K. The relationship between tumor budding and patient survival in breast cancer. Clin Pathol. 2024;17:2632010X241235543. https://doi.org/10.1177/2632010X241235543
205. Wu J, Fan D, Shao Z, Xu B, Ren G, Jiang Z, et al. CACA guidelines for holistic integrative management of breast cancer. Holist Integr Oncol. 2022;1(1):7. https://doi.org/10.1007/s44178-022-00007-8
206. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12-49. https://doi.org/10.3322/caac.21820
207. Xu Y, Gong M, Wang Y, Yang Y, Liu S, Zeng Q. Global trends and forecasts of breast cancer incidence and deaths. Sci Data. 2023;10(1):334. https://doi.org/10.1038/s41597-023-02253-5
208. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263. https://doi.org/10.3322/caac.21834
209. Luo L, Chen Y, Ma Q, Huang Y, Xu L, Shu K, et al. Ginger volatile oil inhibits the growth of MDA-MB-231 in the bisphenol A environment by altering gut microbial diversity. Heliyon. 2024;10(2):e24388. https://doi.org/10.1016/j.heliyon.2024.e24388
210. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271-285. https://doi.org/10.1038/nrc.2017.13
211. Smith A, Pierre JF, Makowski L, Tolley E, Lyn-Cook B, Lu L, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of Non-Hispanic Black and Non-Hispanic White women. Sci Rep. 2019;9(1):11940. https://doi.org/10.1038/s41598-019-48348-1
212. Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS One. 2017;12(11):e0188873. https://doi.org/10.1371/journal.pone.0188873
213. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973-980. https://doi.org/10.1126/science.aay9189
214. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. 2016;6:30751. https://doi.org/10.1038/srep30751
215. Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9(1):e83744. https://doi.org/10.1371/journal.pone.0083744
216. Meng Z, Ye Z, Zhu P, Zhu J, Fang S, Qiu T, et al. New developments and opportunities of microbiota in treating breast cancers. Front Microbiol. 2022;13:818793. https://doi.org/10.3389/fmicb.2022.818793
217. Thu MS, Chotirosniramit K, Nopsopon T, Hirankarn N, Pongpirul K. Human gut, breast, and oral microbiome in breast cancer: a systematic review and meta-analysis. Front Oncol. 2023;13:1144021. https://doi.org/10.3389/fonc.2023.1144021
218. Parida S, Sharma D. The microbiome–estrogen connection and breast cancer risk. Cells. 2019;8(12):1642. https://doi.org/10.3390/cells8121642
219. Kwa M, Plottel CS, Blaser MJ, Adams S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst. 2016;108(8):djw029. https://doi.org/10.1093/jnci/djw029
220. Bernardo G, Le Noci V, Di Modica M, Montanari E, Triulzi T, Pupa SM, et al. The emerging role of the microbiota in breast cancer progression. Cells. 2023;12(15):1945. https://doi.org/10.3390/cells12151945
221. Virtanen V, Paunu K, Ahlskog JK, Varnai R, Sipeky C, Sundvall M. PARP inhibitors in prostate cancer—the preclinical rationale and current clinical development. Genes (Basel). 2019;10(8):565. https://doi.org/10.3390/genes10080565
222. Testa U, Castelli G, Pelosi E. Cellular and molecular mechanisms underlying prostate cancer development: therapeutic implications. Medicines (Basel). 2019;6(3):82. https://doi.org/10.3390/medicines6030082
223. Kopp W. How Western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab Syndr Obes. 2019;12:2221-2236. https://doi.org/10.2147/DMSO.S216791
224. Massari F, Mollica V, Di Nunno V, Gatto L, Santoni M, Scarpelli M, et al. The human microbiota and prostate cancer: friend or foe? Cancers (Basel). 2019;11(4):459. https://doi.org/10.3390/cancers11040459
225. Porter CM, Shrestha E, Peiffer LB, Sfanos KS. The microbiome in prostate inflammation and prostate cancer. Prostate Cancer Prostatic Dis. 2018;21(3):345-354. https://doi.org/10.1038/s41391-018-0041-1
226. Liss MA, White JR, Goros M, Gelfond J, Leach R, Johnson-Pais T, et al. Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur Urol. 2018;74(5):575-582. https://doi.org/10.1016/j.eururo.2018.06.033
227. Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology. 2018;111:122-128. https://doi.org/10.1016/j.urology.2017.08.039
228. Matsushita M, Fujita K, Hatano K, De Velasco MA, Uemura H, Nonomura N. Connecting the dots between the gut–IGF-1–prostate axis: a role of IGF-1 in prostate carcinogenesis. Front Endocrinol (Lausanne). 2022;13:852382. https://doi.org/10.3389/fendo.2022.852382
229. Fujita K, Nonomura N. Role of androgen receptor in prostate cancer: a review. World J Mens Health. 2019;37(3):288-295. https://doi.org/10.5534/wjmh.180040
230. Matsushita M, Fujita K, Motooka D, Hatano K, Hata J, Nishimoto M, et al. Firmicutes in gut microbiota correlate with blood testosterone levels in elderly men. World J Mens Health. 2022;40(3):517-525. https://doi.org/10.5534/wjmh.210190
231. Soheili M, Keyvani H, Soheili M, Nasseri S. Human papillomavirus: a review of epidemiology, carcinogenesis, diagnostic methods, and treatment of all HPV-related cancers. Med J Islam Repub Iran. 2021;35:65. https://doi.org/10.47176/mjiri.35.65
232. Smith JS, Melendy A, Rana RK, Pimenta JM. Age-specific prevalence of infection with human papillomavirus in females: a global review. J Adolesc Health. 2008;43(4 Suppl):S5-S25. https://doi.org/10.1016/j.jadohealth.2008.07.009
233. Watson M, Saraiya M, Benard V, Coughlin SS, Flowers L, Cokkinides V, et al. Burden of cervical cancer in the United States, 1998–2003. Cancer. 2008;113(10 Suppl):2855-2864. https://doi.org/10.1002/cncr.23756
234. Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210(11):1723-1733. https://doi.org/10.1093/infdis/jiu330
235. Laniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynecological cancer development, prevention, and therapy. Nat Rev Urol. 2020;17(4):232-250. https://doi.org/10.1038/s41585-020-0286-z
236. Makker V, MacKay H, Ray-Coquard I, Levine DA, Westin SN, Aoki D, et al. Endometrial cancer. Nat Rev Dis Primers. 2021;7(1):88. https://doi.org/10.1038/s41572-021-00324-8
237. Walther-António MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8(1):122. https://doi.org/10.1186/s13073-016-0368-y
238. Caselli E, Soffritti I, D’Accolti M, Piva I, Greco P, Bonaccorsi G. Atopobium vaginae and Porphyromonas somerae induce proinflammatory cytokine expression in endometrial cells: a possible implication for endometrial cancer? Cancer Manag Res. 2019;11:8571-8575. https://doi.org/10.2147/CMAR.S217362
239. Smolarz B, Biernacka K, Lukasiewicz H, Samulak D, Piekarska E, Romanowicz H, et al. Ovarian cancer - epidemiology, classification, pathogenesis, treatment, and estrogen receptor’ molecular backgrounds. Int J Mol Sci. 2025;26(10):4611. https://doi.org/10.3390/ijms26104611
240. Zhang M, Mo J, Huang W, Bao Y, Luo X, Yuan L. The ovarian cancer-associated microbiome contributes to the tumor’s inflammatory microenvironment. Front Cell Infect Microbiol. 2024;14:1440742. https://doi.org/10.3389/fcimb.2024.1440742
241. Wang Q, Zhao L, Han L, Fu G, Tuo X, Ma S, et al. Differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J Ovarian Res. 2020;13(1):8. https://doi.org/10.1186/s13048-019-0603-4
242. Yu B, Liu C, Proll SC, Manhardt E, Liang S, Srinivasan S, et al. Identification of fallopian tube microbiota and its association with ovarian cancer. eLife. 2024;12:RP89830. https://doi.org/10.7554/eLife.89830
243. Zhou B, Sun C, Huang J, Xia M, Guo E, Li N, et al. Biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. 2019;9(1):1691. https://doi.org/10.1038/s41598-018-38031-2
244. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, et al. The ovarian cancer oncobiome. Oncotarget. 2017;8(22):36225-36245. https://doi.org/10.18632/oncotarget.16717
245. Blanco JR, Del Campo R, Avendaño-Ortiz J, Laguna-Olmos M, Carnero A. The role of microbiota in ovarian cancer: implications for treatment response and therapeutic strategies. Cells. 2025;14(22):1813. https://doi.org/doi:10.3390/cells14221813
246. Huang Q, Wei X, Li W, Ma Y, Chen G, Zhao L, et al. Endogenous Propionibacterium acnes promotes ovarian cancer progression via regulation of the Hedgehog signaling pathway. Cancers (Basel). 2022;14(21):5178. https://doi.org/10.3390/cancers14215178
247. Kawahara N, Yamanaka S, Nishikawa K, Matsuoka M, Maehana T, Kawaguchi R, et al. Endogenous microbacteria can contribute to ovarian carcinogenesis by reducing iron concentration in cysts: a pilot study. Microorganisms. 2024;12(3):538. https://doi.org/10.3390/microorganisms12030538
248. McFarland LV. From yaks to yogurt: the history, development, and current use of probiotics. Clin Infect Dis. 2015;60(Suppl 2):S85-S90. https://doi.org/10.1093/cid/civ054
249. O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol. 2017;2:17057. https://doi.org/10.1038/nmicrobiol.2017.57
250. Shen H, Zhao Z, Zhao Z, Chen Y, Zhang L. Native and engineered probiotics: promising agents against related systemic and intestinal diseases. Int J Mol Sci. 2022;23(2):594. https://doi.org/10.3390/ijms23020594
251. Mahdavi M, Laforest-Lapointe I, Massé E. Preventing colorectal cancer through prebiotics. Microorganisms. 2021;9(6):1325. https://doi.org/10.3390/microorganisms9061325
252. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8(3):92. https://doi.org/10.3390/foods8030092
253. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. 2019;234(10):17127-17143. https://doi.org/10.1002/jcp.28473
254. Zhang M, Liu C, Tu J, Tang M, Ashrafizadeh M, Nabavi N, et al. Advances in cancer immunotherapy: historical perspectives, current developments, and future directions. Mol Cancer. 2025;24:136. https://doi.org/10.1186/s12943-025-02305-x
255. Hua D, Yang Q, Li X, Zhou X, Kang Y, Zhao Y, et al. The combination of Clostridium butyricum and Akkermansia muciniphila mitigates DSS-induced colitis and attenuates colitis-associated tumorigenesis by modulating gut microbiota and reducing CD8(+) T cells in mice. mSystems. 2025;10(2):e01567-24. https://doi.org/10.1128/msystems.01567-24
256. Saeed M, Shoaib A, Kandimalla R, Javed S, Almatroudi A, Gupta R, et al. Microbe-based therapies for colorectal cancer: advantages and limitations. Semin Cancer Biol. 2022;86(Pt 3):652-665. https://doi.org/10.1016/j.semcancer.2021.05.018
257. Gan BK, Rullah K, Yong CY, Ho KL, Omar AR, Alitheen NB, et al. Targeted delivery of 5-fluorouracil-1-acetic acid (5-FA) to cancer cells overexpressing epithelial growth factor receptor (EGFR) using virus-like nanoparticles. Sci Rep. 2020;10(1):16867. https://doi.org/10.1038/s41598-020-73967-4
258. Montassier E, Batard E, Massart S, Gastinne T, Carton T, Caillon J, et al. 16S rRNA gene pyrosequencing reveals a shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol. 2014;67(3):690-699. https://doi.org/10.1007/s00248-013-0355-4
259. van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, et al. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49(2):262-270. https://doi.org/10.1086/599346
260. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510-1519. https://doi.org/10.1136/gutjnl-2019-320204
261. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356-365. https://doi.org/10.1038/nrgastro.2017.20
262. Schwan A, Sjölin S, Trottestam U, Aronsson B. Relapsing Clostridium difficile enterocolitis cured by rectal infusion of homologous faeces. Lancet. 1983;2(8354):845. https://doi.org/10.1016/S0140-6736(83)90753-5
263. Yang R, Chen Z, Cai J. Fecal microbiota transplantation: emerging applications in autoimmune diseases. J Autoimmun. 2023;141:103038. https://doi.org/10.1016/j.jaut.2023.103038
264. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13(9):508-516. https://doi.org/10.1038/nrgastro.2016.98
265. Chang CW, Lee HC, Li LH, Chiang Chiau JS, Wang TE, Chuang WH, et al. Fecal microbiota transplantation prevents intestinal injury, upregulation of Toll-like receptors, and 5-fluorouracil/oxaliplatin-induced toxicity in colorectal cancer. Int J Mol Sci. 2020;21(2):386. https://doi.org/10.3390/ijms21020386
266. Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, Iqbal TH. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther. 2017;46(5):479-493. https://doi.org/10.1111/apt.14201
Declarations
Funding Statement: This work was supported by grants from the National Natural Science Foundation of China (No. 82474506), Shanghai Municipal District-Level General (Specialized) Hospitals’ TCM-WM Integration Specialty Capacity Enhancement Project (No. QJZXYJK-202402), The Key Discipline Development Project of the Shanghai Municipal Health System (No. 2024ZDXK0047), and the “Visiting Renowned Institutions and Seeking Guidance from Distinguished Mentors” Talent Cultivation Program of Shanghai University of Traditional Chinese Medicine (No. 080).
Declaration of Competing Interest: The authors declare no financial or personal relationships with other individuals or organizations that could inappropriately influence or bias the content of this work. All authors have read the final version of the manuscript and confirm that there are no competing interests.
Consent for publication: All authors have approved the final version of the manuscript.
Use of Artificial Intelligence (AI) Disclosure: This article was written by human contributors. Artificial intelligence-based tools were used to improve grammar, language, and readability, without affecting the scientific content, data interpretation, or conclusions of the article. The authors reviewed and verified all content to ensure its accuracy and integrity.
Dual Use Research of Concern (DURC): “The authors confirm that this research does not constitute DURC as defined by the U.S. Government Policy for Oversight of Life Sciences DURC.”
Data Availability Statements (DAS): “No datasets were generated or analyzed in the current study.”
Ethics approval and consent to participate: Not applicable, as this study did not involve the conduct of research.
Authors’ affiliations
1. Department of Pelvic Floor Center, Shanghai Fifth People's Hospital Affiliated to Fudan University, Minhang District Pelvic Floor Center, Shanghai 200240, PR China
2. Institute of Chinese Traditional Surgery, Longhua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai 200032, China.
3. Department of Urology, Longhua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200032, China.
*These authors contributed equally to this article.
# Corresponding authors: Jianfeng Yang Email: 0156333@163.com; Sheng Liu Email: lslhtcm@163.com; Chen Lei - Email: joe8989@163.com
CRediT authorship contribution statement: WY, YS, and JD; investigation, YW and HT; writing the original draft, WY, YS, and JD; visualization, CL, WY, and JD; funding acquisition, YW, SL, and JY; supervision, CL, SL, and JY. All authors have read and agreed to the published version of the manuscript. All authors contributed to the work and approved the final version of the manuscript.
ORCID ID
Sheng Liu: https://orcid.org/0000-0001-8203-9096
How to cite
Wang Y, Shi Y, Duan J, Tang H, Chen L, Liu S, et al. The Cancer Microbiome: Mechanistic and Translational Insights into Oncogenesis and Therapy. Cancer Biome and Targeted Therapy. Published online 2025 Dec.